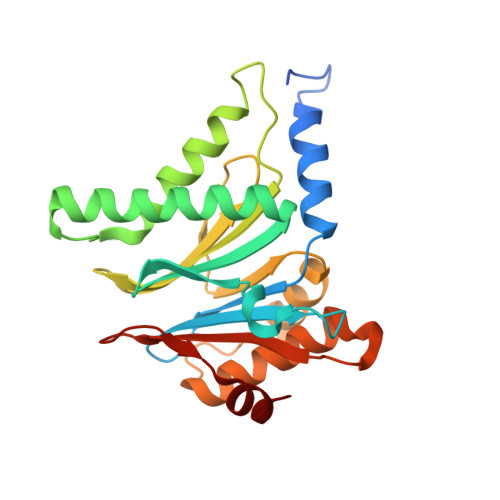
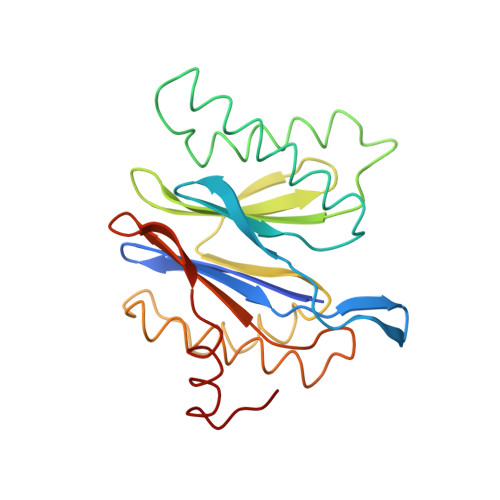
Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution.
Lowe, J., Stock, D., Jap, B., Zwickl, P., Baumeister, W., Huber, R.(1995) Science 268: 533-539
- PubMed: 7725097
- DOI: https://doi.org/10.1126/science.7725097
- Primary Citation of Related Structures:
1PMA - PubMed Abstract:
The three-dimensional structure of the proteasome from the archaebacterium Thermoplasma acidophilum has been elucidated by x-ray crystallographic analysis by means of isomorphous replacement and cyclic averaging. The atomic model was built and refined to a crystallographic R factor of 22.1 percent. The 673-kilodalton protease complex consists of 14 copies of two different subunits, alpha and beta, forming a barrel-shaped structure of four stacked rings. The two inner rings consist of seven beta subunits each, and the two outer rings consist of seven alpha subunits each. A narrow channel controls access to the three inner compartments. The alpha 7 beta 7 beta 7 alpha 7 subunit assembly has 72-point group symmetry. The structures of the alpha and beta subunits are similar, consisting of a core of two antiparallel beta sheets that is flanked by alpha helices on both sides. The binding of a peptide aldehyde inhibitor marks the active site in the central cavity at the amino termini of the beta subunits and suggests a novel proteolytic mechanism.
- Max-Planck-Institut für Biochemie, Abteilung für Strukturforschung, Martinsried, Germany.
Organizational Affiliation: