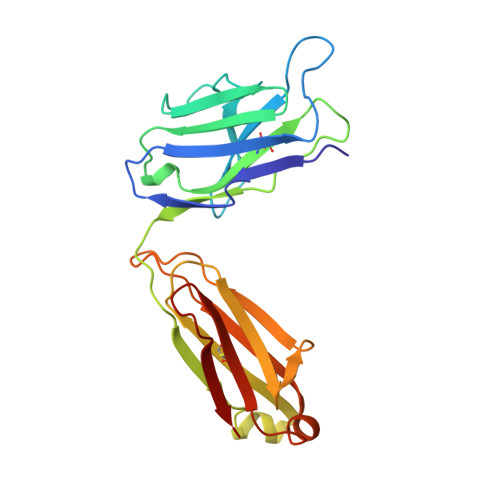
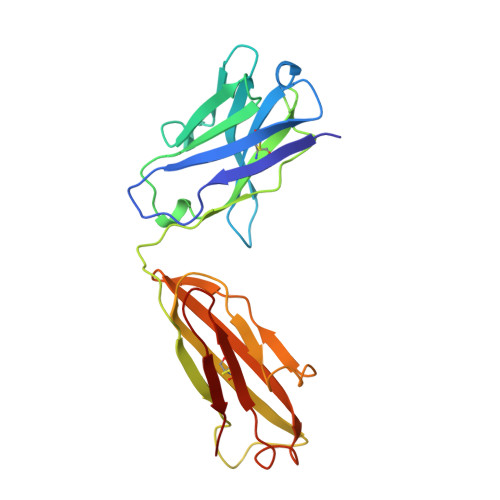
Evidence for the extended helical nature of polysaccharide epitopes. The 2.8 A resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for alpha-(2-->8)-polysialic acid.
Evans, S.V., Sigurskjold, B.W., Jennings, H.J., Brisson, J.R., To, R., Tse, W.C., Altman, E., Frosch, M., Weisgerber, C., Kratzin, H.D., Klebert, S., Vaesen, M., Bitter-Suermann, D., Rose, D.R., Young, N.M., Bundle, D.R.(1995) Biochemistry 34: 6737-6744
- PubMed: 7538787
- DOI: https://doi.org/10.1021/bi00020a019
- Primary Citation of Related Structures:
1PLG - PubMed Abstract:
The antigen binding fragment from an IgG2a kappa murine monoclonal antibody with specificity for alpha-(2-->8)-linked sialic acid polymers has been prepared and crystallized in the absence of hapten. Crystals were grown by vapor diffusion equilibrium with 16-18% polyethylene glycol 4000 solutions. The structure was solved by molecular replacement methods and refined to a conventional R factor of 0.164 for data to 2.8 A. The binding site is observed to display a shape and distribution of charges that is complementary to that of the predicted conformation of the oligosaccharide epitope. A thermodynamic description of ligand binding has been compiled for oligosaccharides ranging in length from 9 to 41 residues, and the data for the largest ligand has been used in a novel way to estimate the size of the antigen binding site. A model of antigen binding is presented that satisfies this thermodynamic data, as well as a previously reported requirement of conformational specificity of the oligosaccharide. X-ray crystallographic and thermodynamic evidence are consistent with a binding site that accommodates at least eight sialic acid residues.
- Institute for Biological Sciences, National Research Council of Canada, Ottawa.
Organizational Affiliation: