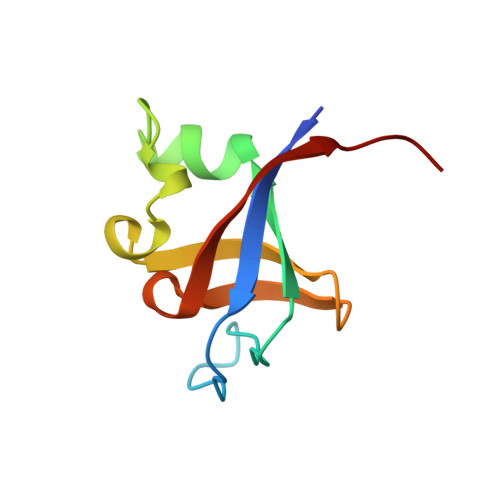
Crystal structure of P13K SH3 domain at 20 angstroms resolution.
Liang, J., Chen, J.K., Schreiber, S.T., Clardy, J.(1996) J Mol Biology 257: 632-643
- PubMed: 8648629
- DOI: https://doi.org/10.1006/jmbi.1996.0190
- Primary Citation of Related Structures:
1PHT - PubMed Abstract:
The P13K SH3 domain, residues 1 to 85 of the P1-3 kinase p85 subunit, has been characterized by X-ray diffraction. Crystals belonging to space group P4(3)2(1)2 diffract to 2.0 angstroms resolution and the structure was phased by single isomorphous replacement and anomalous scattering (SIRAS). As expected, the domain is a compact beta barrel with an over-all confirmation very similar to the independently determined NMR structures. The X-ray structure illuminates a discrepancy between the two NMR structures on the conformation of the loop region unique to P13K SH3. Furthermore, the ligand binding pockets of P13K SH3 domain are occupied by amino acid residues from symmetry-related P13K SH3 molecules: the C-terminal residues I(82) SPP of one and R18 of another. The interaction modes clearly resemble those observed for the P13K SH3 domain complexed with the synthetic peptide RLP1, a class 1 ligand, although there are significant differences. The solid-state interactions suggest a model of protein-protein aggregation that could be mediated by SH3 domains.
- Department of Chemistry, Baker Laboratory. Cornell University, Ithaca, NY 14853-1301, USA.
Organizational Affiliation: