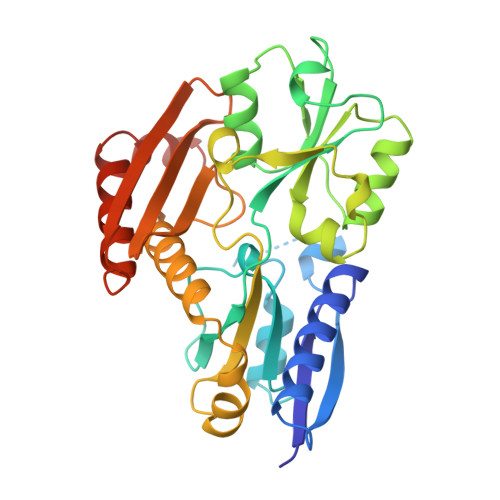
Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site.
Louie, G.V., Brownlie, P.D., Lambert, R., Cooper, J.B., Blundell, T.L., Wood, S.P., Warren, M.J., Woodcock, S.C., Jordan, P.M.(1992) Nature 359: 33-39
- PubMed: 1522882
- DOI: https://doi.org/10.1038/359033a0
- Primary Citation of Related Structures:
1PDA - PubMed Abstract:
The three-domain structure of porphobilinogen deaminase, a key enzyme in the biosynthetic pathway of tetrapyrroles, has been defined by X-ray analysis at 1.9 A resolution. Two of the domains structurally resemble the transferrins and periplasmic binding proteins. The dipyrromethane cofactor is covalently linked to domain 3 but is bound by extensive salt-bridges and hydrogen-bonds within the cleft between domains 1 and 2, at a position corresponding to the binding sites for small-molecule ligands in the analogous proteins. The X-ray structure and results from site-directed mutagenesis provide evidence for a single catalytic site. Interdomain flexibility may aid elongation of the polypyrrole product in the active-site cleft of the enzyme.
- Department of Crystallography, Birkbeck College, University of London, UK.
Organizational Affiliation: