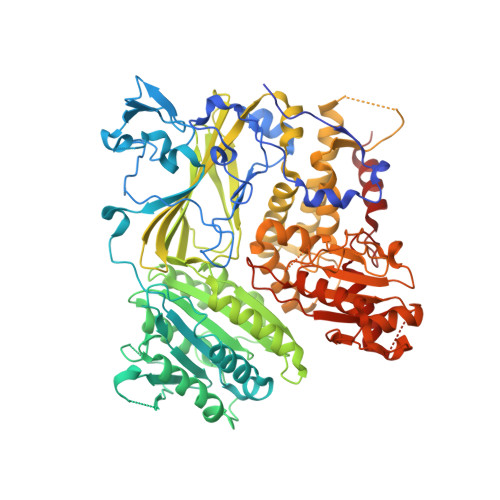
SNARE selectivity of the COPII coat.
Mossessova, E., Bickford, L.C., Goldberg, J.(2003) Cell 114: 483-495
- PubMed: 12941276
- DOI: https://doi.org/10.1016/s0092-8674(03)00608-1
- Primary Citation of Related Structures:
1PCX, 1PD0, 1PD1 - PubMed Abstract:
The COPII coat buds transport vesicles from the endoplasmic reticulum that incorporate cargo and SNARE molecules. Here, we show that recognition of the ER-Golgi SNAREs Bet1, Sed5, and Sec22 occurs through three binding sites on the Sec23/24 subcomplex of yeast COPII. The A site binds to the YNNSNPF motif of Sed5. The B site binds to Lxx-L/M-E sequences present in both the Bet1 and Sed5 molecules, as well as to the DxE cargo-sorting signal. A third, spatially distinct site binds to Sec22. COPII selects the free v-SNARE form of Bet1 because the LxxLE sequence is sequestered in the four-helix bundle of the v-/t-SNARE complex. COPII favors Sed5 within the Sed5/Bos1/Sec22 t-SNARE complex because t-SNARE assembly removes autoinhibitory contacts to expose the YNNSNPF motif. The COPII coat seems to be a specific conductor of the fusogenic forms of these SNAREs, suggesting how vesicle fusion specificity may be programmed during budding.
- Howard Hughes Medical Institute and the Cellular Biochemistry and Biophysics Program, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, USA.
Organizational Affiliation: