Helix-stabilized cyclic peptides as selective inhibitors of steroid receptor-coactivator interactions
Leduc, A.M., Trent, J.O., Wittliff, J.L., Bramlett, K.S., Briggs, S.L., Chirgadze, N.Y., Wang, Y., Burris, T.P., Spatola, A.F.(2003) Proc Natl Acad Sci U S A 100: 11273-11278
- PubMed: 13679575
- DOI: https://doi.org/10.1073/pnas.1934759100
- Primary Citation of Related Structures:
1PCG - PubMed Abstract:
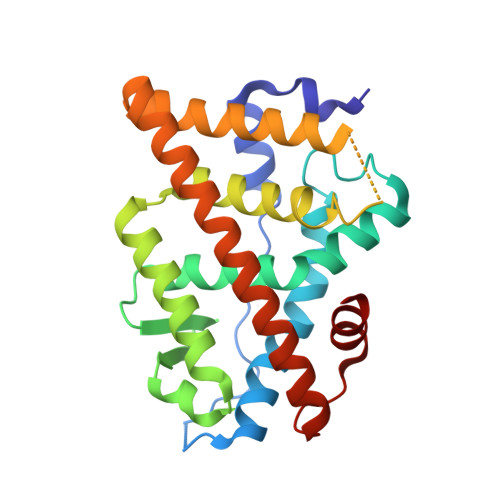
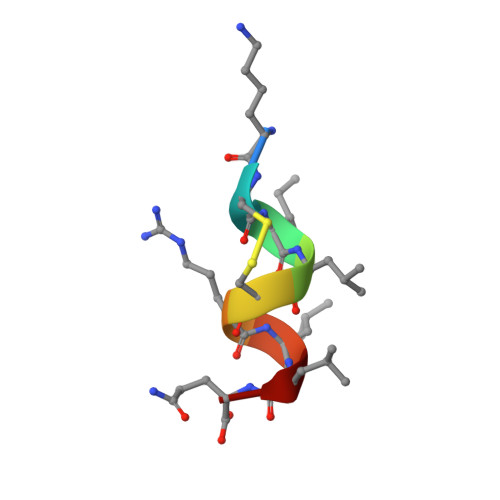
The interaction between nuclear receptors and coactivators provides an arena for testing whether protein-protein interactions may be inhibited by small molecule drug candidates. We provide evidence that a short cyclic peptide, containing a copy of the LXXLL nuclear receptor box pentapeptide, binds tightly and selectively to estrogen receptor alpha. Furthermore, as shown by x-ray analysis, the disulfide-bridged nonapeptide, nonhelical in aqueous solutions, is able to adopt a quasihelical conformer while binding to the groove created by ligand attachment to estrogen receptor alpha. An i, i+3 linked analog, H-Lys-cyclo(d-Cys-Ile-Leu-Cys)-Arg-Leu-Leu-Gln-NH2 (peptidomimetic estrogen receptor modulator 1), binds with a Ki of 25 nM, significantly better than an i, i+4 bridged cyclic amide, as predicted by molecular modeling design criteria. The induction of helical character, effective binding, and receptor selectivity exhibited by this peptide analog provide strong support for this strategy. The stabilization of minimalist surface motifs may prove useful for the control of other macromolecular assemblies, especially when an amphiphilic helix is crucial for the strong binding interaction between two proteins.
- Department of Chemistry, University of Louisville, Louisville, KY 40292, USA.
Organizational Affiliation: