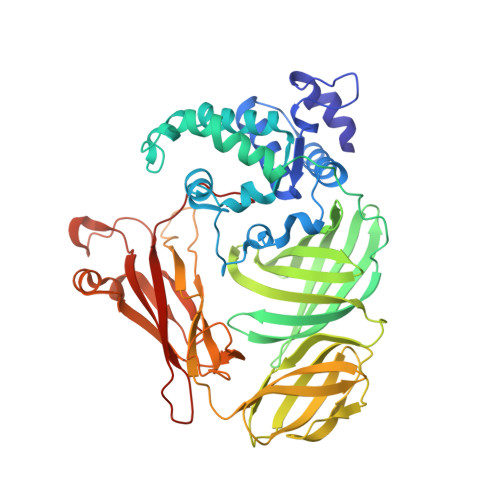
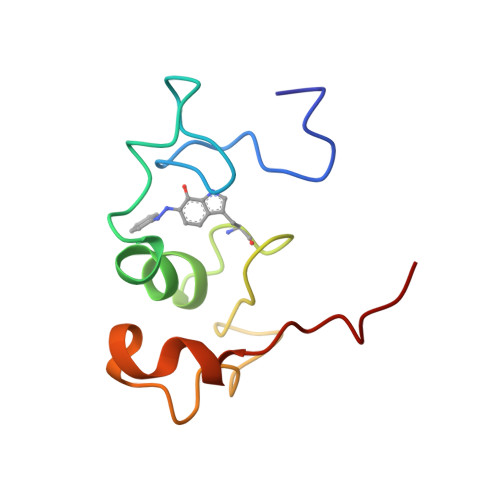
Structure of the phenylhydrazine adduct of the quinohemoprotein amine dehydrogenase from Paracoccus denitrificans at 1.7 A resolution.
Datta, S., Ikeda, T., Kano, K., Mathews, F.S.(2003) Acta Crystallogr D Biol Crystallogr 59: 1551-1556
- PubMed: 12925784
- DOI: https://doi.org/10.1107/s090744490301429x
- Primary Citation of Related Structures:
1PBY - PubMed Abstract:
The 109 kDa quinohemoprotein amine dehydrogenase (QHNDH) from Paracoccus denitrificans contains a novel redox cofactor, cysteine tryptophylquinone (CTQ). This cofactor is derived from a pair of gene-encoded amino acids by post-translational modification and was previously identified and characterized within an 82-residue subunit by chemical methods and crystallographic analysis at 2.05 A resolution. It contains an orthoquinone moiety bound to the indole ring and catalyzes the oxidation of aliphatic and aromatic amines through formation of a Schiff-base intermediate involving one of the quinone O atoms. This paper reports the structural analysis of the complex of QHNDH with the enzyme inhibitor phenylhydrazine determined at 1.70 A resolution. The phenylhydrazone product is attached to the C6 position, identifying the O6 atom of CTQ as the site of Schiff-base formation as postulated by analogy to another amine-oxidizing enzyme, methylamine dehydrogenase. Furthermore, the inner N atom closest to the phenyl ring of phenylhydrazine forms a hydrogen bond to gammaAsp33 in the complex, lending support to the hypothesis that this residue serves as the active-site base for proton abstraction during catalysis.
- Washington University School of Medicine, St Louis, MO 63110, USA.
Organizational Affiliation: