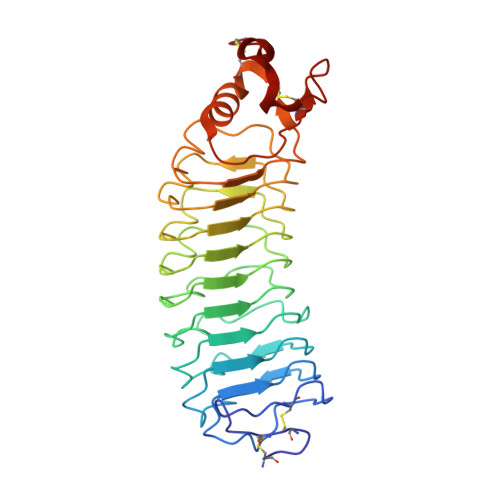
Structure and axon outgrowth inhibitor binding of the Nogo-66 receptor and related proteins
Barton, W.A., Liu, B.P., Tzvetkova, D., Jeffrey, P.D., Fournier, A.E., Sah, D., Cate, R., Strittmatter, S.M., Nikolov, D.B.(2003) EMBO J 22: 3291-3302
- PubMed: 12839991
- DOI: https://doi.org/10.1093/emboj/cdg325
- Primary Citation of Related Structures:
1P8T - PubMed Abstract:
The myelin-derived proteins Nogo, MAG and OMgp limit axonal regeneration after injury of the spinal cord and brain. These cell-surface proteins signal through multi-subunit neuronal receptors that contain a common ligand-binding glycosylphosphatidylinositol-anchored subunit termed the Nogo-66 receptor (NgR). By deletion analysis, we show that the binding of soluble fragments of Nogo, MAG and NgR to cell-surface NgR requires the entire leucine-rich repeat (LRR) region of NgR, but not other portions of the protein. Despite sharing extensive sequence similarity with NgR, two related proteins, NgR2 and NgR3, which we have identified, do not bind Nogo, MAG, OMgp or NgR. To investigate NgR specificity and multi-ligand binding, we determined the crystal structure of the biologically active ligand-binding soluble ectodomain of NgR. The molecule is banana shaped with elongation and curvature arising from eight LRRs flanked by an N-terminal cap and a small C-terminal subdomain. The NgR structure analysis, as well as a comparison of NgR surface residues not conserved in NgR2 and NgR3, identifies potential protein interaction sites important in the assembly of a functional signaling complex.
- Cellular Biochemistry and Biophysics Program, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, USA.
Organizational Affiliation: