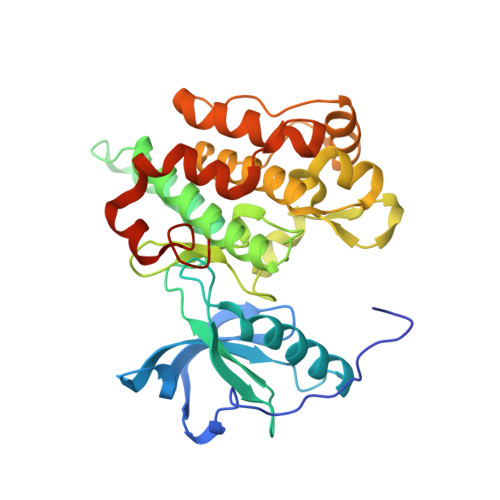
Structure of apo, unactivated insulin-like growth factor-1 receptor kinase at 1.5 A resolution.
Munshi, S., Hall, D.L., Kornienko, M., Darke, P.L., Kuo, L.C.(2003) Acta Crystallogr D Biol Crystallogr 59: 1725-1730
- PubMed: 14501110
- DOI: https://doi.org/10.1107/s0907444903015415
- Primary Citation of Related Structures:
1P4O - PubMed Abstract:
The crystal structure of the wild-type unactivated kinase domain (IGFRK-0P) of insulin-like growth factor-1 receptor has been reported previously at 2.7 A resolution [Munshi et al. (2002), J. Biol. Chem. 277, 38797-38802]. In order to obtain a high-resolution structure, a number of variants of IGFRK-0P were prepared and screened for crystallization. A double mutant with E1067A and E1069A substitutions within the kinase-insert region resulted in crystals that diffracted to 1.5 A resolution. Overall, the structure of the mutant IGFRK-0P is similar to that of the wild-type IGFRK-0P structure, with the exception of the previously disordered kinase-insert region in the wild type having become fixed. In addition, amino-acid residues 947-952 at the N-terminus are well defined in the mutant structure. The monomeric protein structure is folded into two lobes connected by a hinge region, with the catalytic center situated at the interface of the two lobes. Two molecules of IGFRK-0P in the asymmetric unit are associated as a dimer and two different types of dimers with their ATP-binding clefts either facing towards or away from each other are observed. The current refined model consists of a dimer and 635 water molecules.
- Department of Structural Biology, Merck Research Laboratories, West Point, PA 19486, USA. sanjeev_munshi@merck.com
Organizational Affiliation: