Structure of NFAT Bound to DNA as a Monomer
Stroud, J.C., Chen, L.(2003) J Mol Biology 334: 1009-1022
- PubMed: 14643663
- DOI: https://doi.org/10.1016/j.jmb.2003.09.065
- Primary Citation of Related Structures:
1OWR - PubMed Abstract:
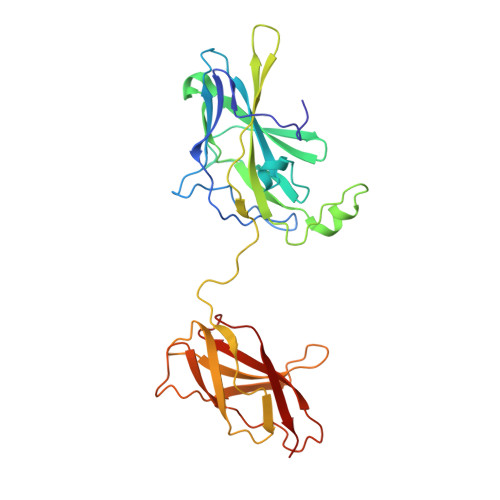
The nuclear factor of activated T cells (NFAT) is a calcium-dependent transcription factor that cooperates with a myriad of partner transcription factors to regulate distinct transcription programs. Transcription activation by NFAT without the cooperation of co-stimulatory signals in lymphocytes can also impose a genetic program of anergy. Although the ternary NFAT1/Fos-Jun/DNA complex has been structurally characterized, how NFAT1 recognizes DNA in the absence of cooperative partners and how such a binary NFAT/DNA complex may lead to the assembly of distinct high-order NFAT transcription complexes are still poorly understood. We have determined the crystal structure of the entire Rel homology region (RHR) of human NFAT1 (NFATc2) bound to DNA as a monomer. We also present footprinting evidence that corroborates the protein-DNA contacts observed in the crystal structure. Our structural and biochemical studies reveal the mechanism by which the monomeric Rel protein NFAT recognizes its cognate DNA site. A remarkable feature of the binary NFAT/DNA complex is the conformational flexibility exhibited by NFAT1 in the four independent copies of the NFAT/DNA complex in the crystal structure, which may reflect a mechanism by which NFAT1 interacts with a variety of protein partners as it mediates disparate biological responses.
- Department of Chemistry and Biochemistry, University of Colorado at Boulder, Campus Box 215, Boulder, CO 80309-0437, USA.
Organizational Affiliation: