Differential thymic selection outcomes stimulated by focal structural alteration in peptide/major histocompatibility complex ligands.
Ghendler, Y., Teng, M.K., Liu, J.H., Witte, T., Liu, J., Kim, K.S., Kern, P., Chang, H.C., Wang, J.H., Reinherz, E.L.(1998) Proc Natl Acad Sci U S A 95: 10061-10066
- PubMed: 9707600
- DOI: https://doi.org/10.1073/pnas.95.17.10061
- Primary Citation of Related Structures:
1OSZ - PubMed Abstract:
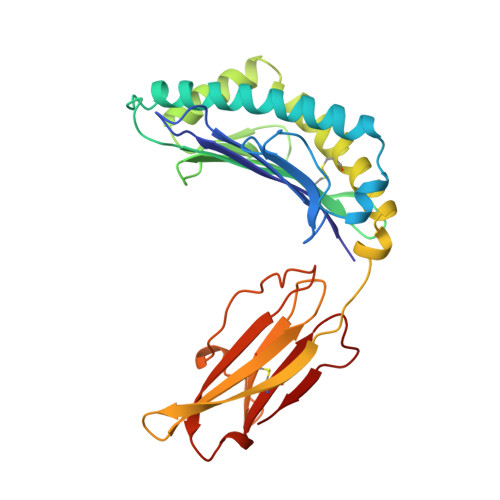
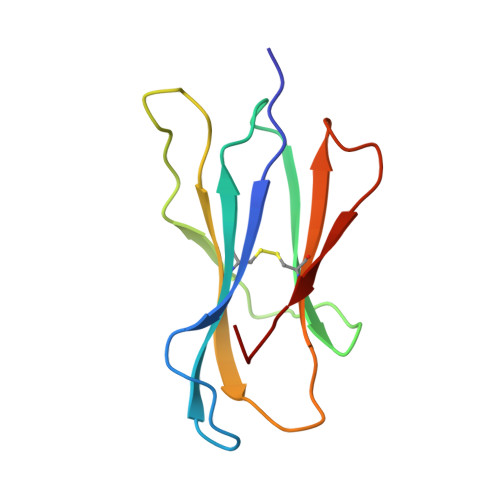
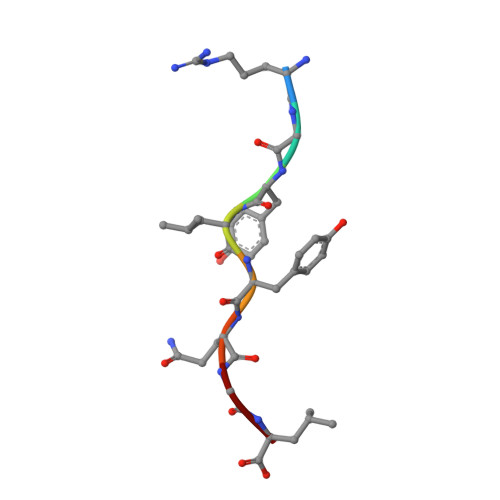
The T lineage repertoire is shaped by T cell receptor (TCR)-dependent positive and negative thymic selection processes. Using TCR-transgenic (N15tg) beta2-microglobulin-deficient (beta2m-/-) RAG-2(-/-) H-2(b) mice specific for the VSV8 (RGYVYQGL) octapeptide bound to Kb, we identified a single weak agonist peptide variant V4L (L4) inducing phenotypic and functional T cell maturation. The cognate VSV8 peptide, in contrast, triggers negative selection. The crystal structure of L4/Kb was determined and refined to 2.1 A for comparison with the VSV8/Kb structure at similar resolution. Aside from changes on the p4 side chain of L4 and the resulting alteration of the exposed Kb Lys-66 side chain, these two structures are essentially identical. Hence, a given TCR recognizes subtle distinctions between highly related ligands, resulting in dramatically different selection outcomes. Based on these finding and the recent structural elucidation of the N15-VSV8/Kb complex, moreover, it appears that the germ-line Valpha repertoire contributes in a significant way to positive selection.
- Laboratory of Immunobiology, Dana-Farber Cancer Institute and Department of Medicine, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: