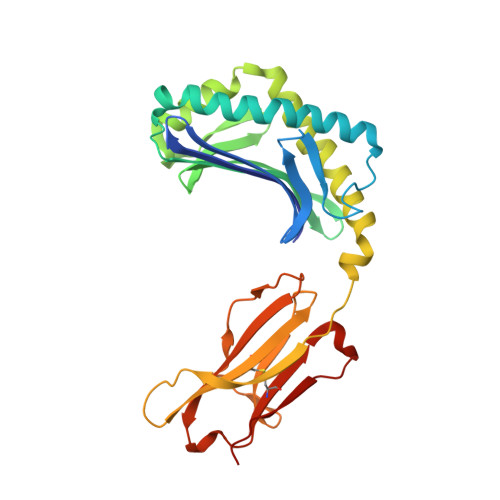
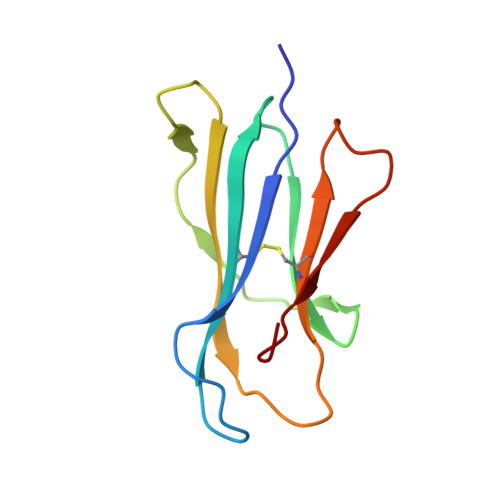
Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A.
Zajonc, D.M., Elsliger, M.A., Teyton, L., Wilson, I.A.(2003) Nat Immunol 4: 808-815
- PubMed: 12833155
- DOI: https://doi.org/10.1038/ni948
- Primary Citation of Related Structures:
1ONQ - PubMed Abstract:
CD1 antigens bind a variety of self and foreign lipid and glycolipid antigens for presentation to CD1-restricted T cell receptors (TCRs). Here we report the crystal structure of human CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. The lipid adopts an S-shaped conformation, with the sphingosine chain completely buried in the A' pocket and the fatty acid chain emerging from the interface of the A' pocket into the more exposed F' pocket. The headgroup is anchored in the A'-F' junction and protrudes into the F' pocket for TCR recognition. Because the A' pocket is narrow with a fixed terminus, it can act as a molecular 'ruler' to select alkyl chains of a particular length.
- Department of Molecular Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, USA.
Organizational Affiliation: