Structural insight into the role of the ribosomal tunnel in cellular regulation
Berisio, R., Schluenzen, F., Harms, J., Bashan, A., Auerbach, T., Baram, D., Yonath, A.(2003) Nat Struct Biol 10: 366-370
- PubMed: 12665853
- DOI: https://doi.org/10.1038/nsb915
- Primary Citation of Related Structures:
1OND - PubMed Abstract:
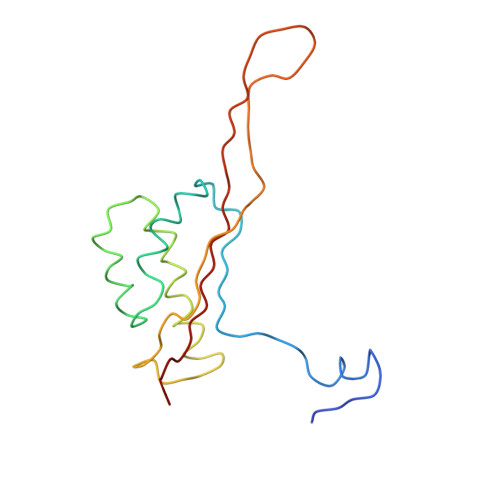
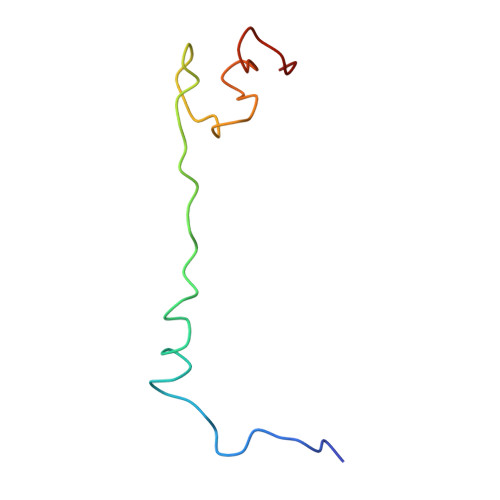
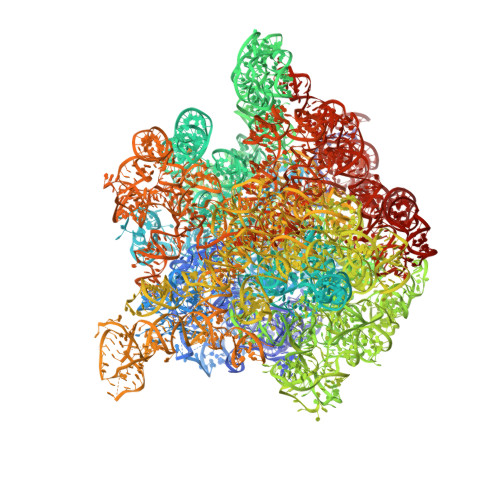
Nascent proteins emerge out of ribosomes through an exit tunnel, which was assumed to be a firmly built passive path. Recent biochemical results, however, indicate that the tunnel plays an active role in sequence-specific gating of nascent chains and in responding to cellular signals. Consistently, modulation of the tunnel shape, caused by the binding of the semi-synthetic macrolide troleandomycin to the large ribosomal subunit from Deinococcus radiodurans, was revealed crystallographically. The results provide insights into the tunnel dynamics at high resolution. Here we show that, in addition to the typical steric blockage of the ribosomal tunnel by macrolides, troleandomycin induces a conformational rearrangement in a wall constituent, protein L22, flipping the tip of its highly conserved beta-hairpin across the tunnel. On the basis of mutations that alleviate elongation arrest, the tunnel motion could be correlated with sequence discrimination and gating, suggesting that specific arrest motifs within nascent chain sequences may induce a similar gating mechanism.
- Max-Planck-Research Unit for Ribosomal Structure, Hamburg, Germany.
Organizational Affiliation: