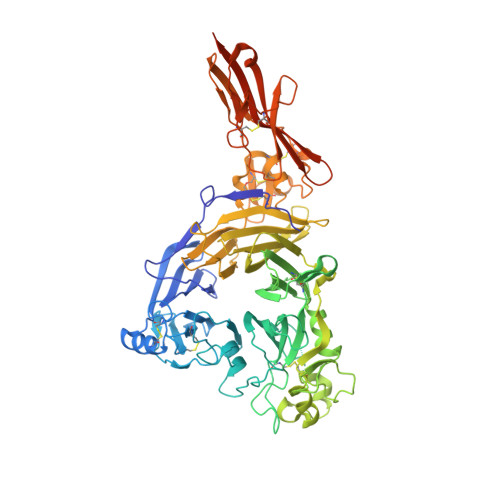
The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Love, C.A., Harlos, K., Mavaddat, N., Davis, S.J., Stuart, D.I., Jones, E.Y., Esnouf, R.M.(2003) Nat Struct Biol 10: 843
- PubMed: 12958590
- DOI: https://doi.org/10.1038/nsb977
- Primary Citation of Related Structures:
1OLZ - PubMed Abstract:
Semaphorins, proteins characterized by an extracellular sema domain, regulate axon guidance, immune function and angiogenesis. The crystal structure of SEMA4D (residues 1-657) shows the sema topology to be a seven-bladed beta-propeller, revealing an unexpected homology with integrins. The sema beta-propeller contains a distinctive 77-residue insertion between beta-strands C and D of blade 5. Blade 7 is followed by a domain common to plexins, semaphorins and integrins (PSI domain), which forms a compact cysteine knot abutting the side of the propeller, and an Ig-like domain. The top face of the beta-propeller presents prominent loops characteristic of semaphorins. In addition to limited contact between the Ig-like domains, the homodimer is stabilized through extensive interactions between the top faces in a sector of the beta-propeller used for heterodimerization in integrins. This face of the propeller also mediates ligand binding in integrins, and functional data for semaphorin-receptor interactions map to the equivalent surface.
- Division of Structural Biology, Henry Wellcome Building for Genomic Medicine, University of Oxford, Roosevelt Drive, Oxford, OX3 7BN, UK.
Organizational Affiliation: