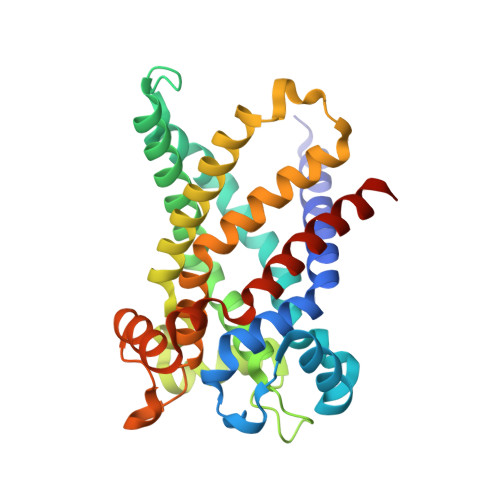
Structure of Mitochondrial Adp/ATP Carrier in Complex with Carboxyatractyloside
Pebay-Peyroula, E., Dahout-Gonzalez, C., Kahn, R., Trezeguet, V., Lauquin, G.J.-M., Brandolin, G.(2003) Nature 426: 39
- PubMed: 14603310
- DOI: https://doi.org/10.1038/nature02056
- Primary Citation of Related Structures:
1OKC - PubMed Abstract:
ATP, the principal energy currency of the cell, fuels most biosynthetic reactions in the cytoplasm by its hydrolysis into ADP and inorganic phosphate. Because resynthesis of ATP occurs in the mitochondrial matrix, ATP is exported into the cytoplasm while ADP is imported into the matrix. The exchange is accomplished by a single protein, the ADP/ATP carrier. Here we have solved the bovine carrier structure at a resolution of 2.2 A by X-ray crystallography in complex with an inhibitor, carboxyatractyloside. Six alpha-helices form a compact transmembrane domain, which, at the surface towards the space between inner and outer mitochondrial membranes, reveals a deep depression. At its bottom, a hexapeptide carrying the signature of nucleotide carriers (RRRMMM) is located. Our structure, together with earlier biochemical results, suggests that transport substrates bind to the bottom of the cavity and that translocation results from a transient transition from a 'pit' to a 'channel' conformation.
- Institut de Biologie Structurale, UMR 5075 CEA-CNRS-Université Joseph Fourier, 41 rue Jules Horowitz, F-38027, Grenoble cedex 1, France. pebay@ibs.fr
Organizational Affiliation: