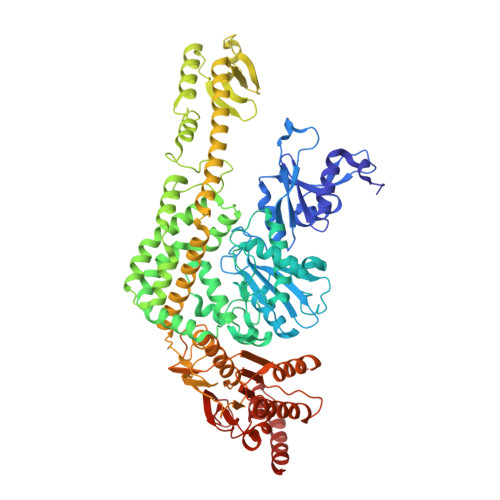
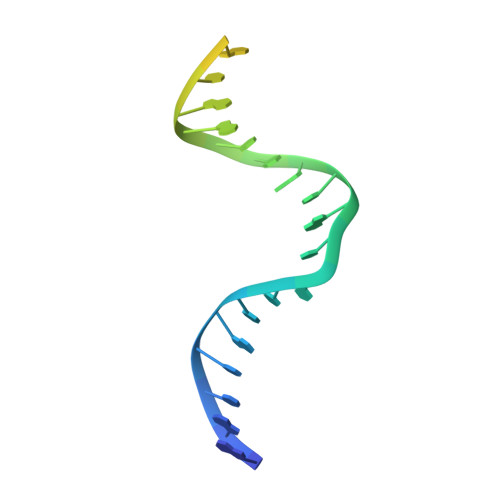
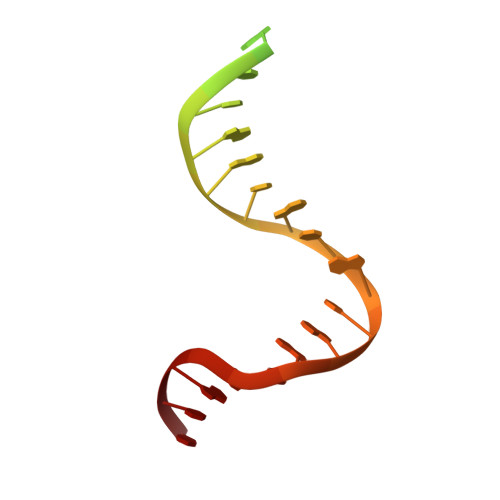
Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Natrajan, G., Lamers, M.H., Enzlin, J.H., Winterwerp, H.H.K., Perrakis, A., Sixma, T.K.(2003) Nucleic Acids Res 31: 4814
- PubMed: 12907723
- DOI: https://doi.org/10.1093/nar/gkg677
- Primary Citation of Related Structures:
1OH5, 1OH6, 1OH7, 1OH8 - PubMed Abstract:
We have refined a series of isomorphous crystal structures of the Escherichia coli DNA mismatch repair enzyme MutS in complex with G:T, A:A, C:A and G:G mismatches and also with a single unpaired thymidine. In all these structures, the DNA is kinked by approximately 60 degrees upon protein binding. Two residues widely conserved in the MutS family are involved in mismatch recognition. The phenylalanine, Phe 36, is seen stacking on one of the mismatched bases. The same base is also seen forming a hydrogen bond to the glutamate Glu 38. This hydrogen bond involves the N7 if the base stacking on Phe 36 is a purine and the N3 if it is a pyrimidine (thymine). Thus, MutS uses a common binding mode to recognize a wide range of mismatches.
- Division of Molecular Carcinogenesis, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX, Amsterdam, The Netherlands.
Organizational Affiliation: