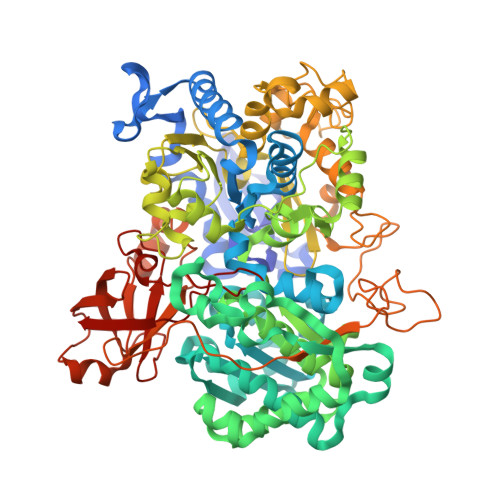
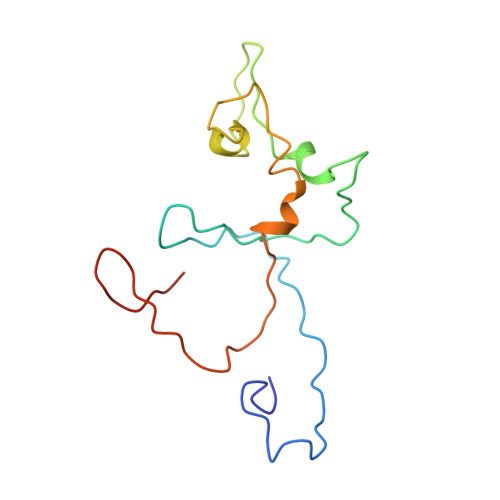
Structural and Redox Plasticity in the Heterodimeric Periplasmic Nitrate Reductase
Arnoux, P., Sabaty, M., Alric, J., Frangioni, B., Guigliarelli, B., Adriano, J.-M., Pignol, D.(2003) Nat Struct Biol 10: 928
- PubMed: 14528294
- DOI: https://doi.org/10.1038/nsb994
- Primary Citation of Related Structures:
1OGY - PubMed Abstract:
The structure of the respiratory nitrate reductase (NapAB) from Rhodobacter sphaeroides, the periplasmic heterodimeric enzyme responsible for the first step in the denitrification process, has been determined at a resolution of 3.2 A. The di-heme electron transfer small subunit NapB binds to the large subunit with heme II in close proximity to the [4Fe-4S] cluster of NapA. A total of 57 residues at the N- and C-terminal extremities of NapB adopt an extended conformation, embracing the NapA subunit and largely contributing to the total area of 5,900 A(2) buried in the complex. Complex formation was studied further by measuring the variation of the redox potentials of all the cofactors upon binding. The marked effects observed are interpreted in light of the three-dimensional structure and depict a plasticity that contributes to an efficient electron transfer in the complex from the heme I of NapB to the molybdenum catalytic site of NapA.
- CEA/Cadarache, DSV, DEVM, Laboratoire de Bioénergétique Cellulaire, 13108 St Paul lez Durance Cedex, France. pascal.arnoux@cea.fr
Organizational Affiliation: