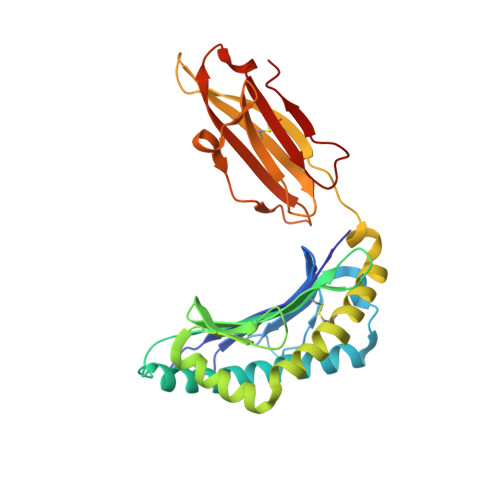
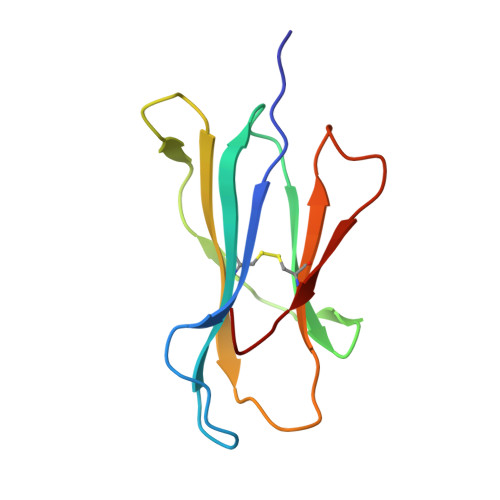
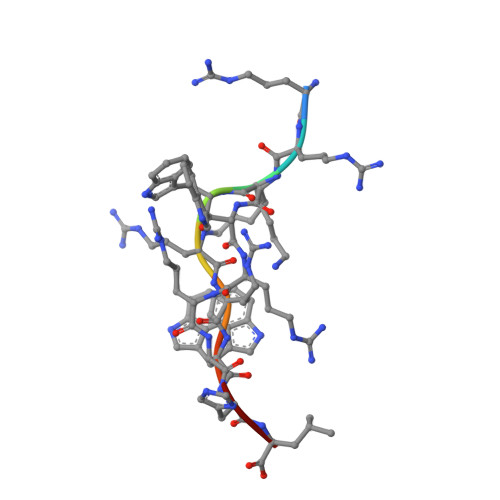
Dual, HLA-B27 subtype-dependent conformation of a self-peptide.
Hulsmeyer, M., Fiorillo, M.T., Bettosini, F., Sorrentino, R., Saenger, W., Ziegler, A., Uchanska-Ziegler, B.(2004) J Exp Medicine 199: 271-281
- PubMed: 14734527
- DOI: https://doi.org/10.1084/jem.20031690
- Primary Citation of Related Structures:
1OF2, 1OGT - PubMed Abstract:
The products of the human leukocyte antigen subtypes HLA-B*2705 and HLA-B*2709 differ only in residue 116 (Asp vs. His) within the peptide binding groove but are differentially associated with the autoimmune disease ankylosing spondylitis (AS); HLA-B*2705 occurs in AS-patients, whereas HLA-B*2709 does not. The subtypes also generate differential T cell repertoires as exemplified by distinct T cell responses against the self-peptide pVIPR (RRKWRRWHL). The crystal structures described here show that pVIPR binds in an unprecedented dual conformation only to HLA-B*2705 molecules. In one binding mode, peptide pArg5 forms a salt bridge to Asp116, connected with drastically different interactions between peptide and heavy chain, contrasting with the second, conventional conformation, which is exclusively found in the case of B*2709. These subtype-dependent differences in pVIPR binding link the emergence of dissimilar T cell repertoires in individuals with HLA-B*2705 or HLA-B*2709 to the buried Asp116/His116 polymorphism and provide novel insights into peptide presentation by major histocompatibility antigens.
- Institut für Kristallographie, Freie Universität Berlin, 14195 Berlin, Germany.
Organizational Affiliation: