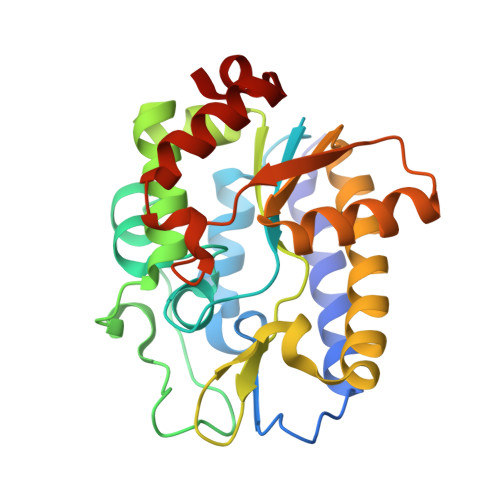
Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase Smug1
Wibley, J.E.A., Waters, T.R., Haushalter, K., Verdine, G.L., Pearl, L.H.(2003) Mol Cell 11: 1647
- PubMed: 12820976
- DOI: https://doi.org/10.1016/s1097-2765(03)00235-1
- Primary Citation of Related Structures:
1OE4, 1OE5, 1OE6 - PubMed Abstract:
Cytosine deamination is a major promutagenic process, generating G:U mismatches that can cause transition mutations if not repaired. Uracil is also introduced into DNA via nonmutagenic incorporation of dUTP during replication. In bacteria, uracil is excised by uracil-DNA glycosylases (UDG) related to E. coli UNG, and UNG homologs are found in mammals and viruses. Ung knockout mice display no increase in mutation frequency due to a second UDG activity, SMUG1, which is specialized for antimutational uracil excision in mammalian cells. Remarkably, SMUG1 also excises the oxidation-damage product 5-hydroxymethyluracil (HmU), but like UNG is inactive against thymine (5-methyluracil), a chemical substructure of HmU. We have solved the crystal structure of SMUG1 complexed with DNA and base-excision products. This structure indicates a more invasive interaction with dsDNA than observed with other UDGs and reveals an elegant water displacement/replacement mechanism that allows SMUG1 to exclude thymine from its active site while accepting HmU.
- Cancer Research UK DNA Repair Enzyme Group, Section of Structural Biology, The Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Road, London SW3 6JB, United Kingdom.
Organizational Affiliation: