Structure of the Oct-3 POU-Homeodomain in Solution, as Determined by Triple Resonance Heteronuclear Multidimensional NMR Spectroscopy
Morita, E.H., Shirakawa, M., Hayashi, F., Imagawa, M., Kyogoku, Y.(1995) Protein Sci 4: 729-739
- PubMed: 7613470
- DOI: https://doi.org/10.1002/pro.5560040412
- Primary Citation of Related Structures:
1OCP - PubMed Abstract:
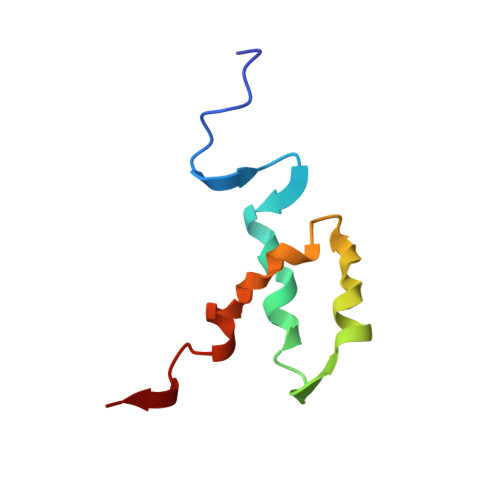
The POU-homeodomain (POUH) forms the bipartite DNA-binding POU domain in association with the POU-specific domain. The 1H, 15N, and 13C magnetic resonances of the 67-amino acid long POUH of mouse Oct-3 have almost completely been assigned, mainly through the combined use of three-dimensional triple resonance NMR methods. Based on the distance and dihedral angle constraints derived from the NMR data, the solution structure of the POUH domain has been calculated by the ab initio simulated annealing method. The average RMS deviation for all backbone heavy atoms of the 20 best calculated structures for residues 9-53 of the total 67 amino acid residues is 0.44 A. The POUH domain consists of three alpha-helices (helix-I, 10-20; helix-II, 28-38; and helix-III, 42-53), and helices-II and -III form a helix-turn-helix motif. In comparison with other classical homeodomains, the folding of the three helices is quite similar. However, the length of helix-III is fairly short. In the complex of the Oct-1 POU domain with an octamer site (Klemm JD, et al., 1994, Cell 77:21-32), the corresponding region is involved in helix-III. The structural difference between these two cases will be discussed.
- Institute for Protein Research, Osaka University, Japan.
Organizational Affiliation: