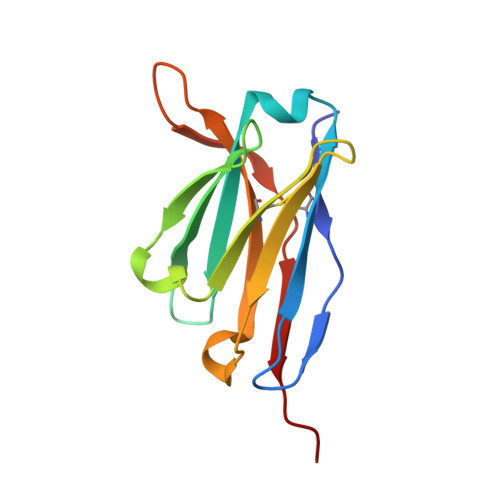
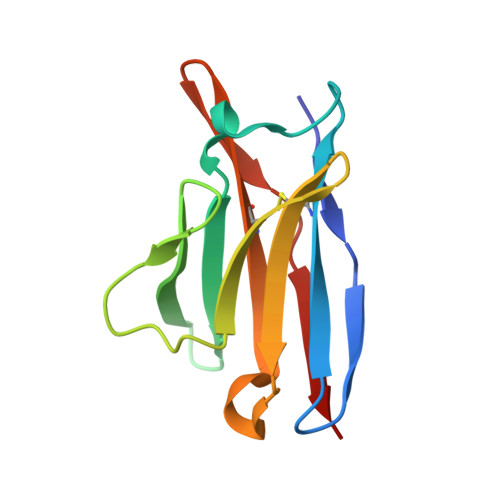
Antibody Multispecificity Mediated by Conformational Diversity
James, L.C., Roversi, P., Tawfik, D.(2003) Science 299: 1362
- PubMed: 12610298
- DOI: https://doi.org/10.1126/science.1079731
- Primary Citation of Related Structures:
1OAQ, 1OAR, 1OAU, 1OAX, 1OAY, 1OAZ, 1OCW - PubMed Abstract:
A single antibody was shown to adopt different binding-site conformations and thereby bind unrelated antigens. Analysis by both x-ray crystallography and pre-steady-state kinetics revealed an equilibrium between different preexisting isomers, one of which possessed a promiscuous, low-affinity binding site for aromatic ligands, including the immunizing hapten. A subsequent induced-fit isomerization led to high-affinity complexes with a deep and narrow binding site. A protein antigen identified by repertoire selection made use of an unrelated antibody isomer with a wide, shallow binding site. Conformational diversity, whereby one sequence adopts multiple structures and multiple functions, can increase the effective size of the antibody repertoire but may also lead to autoimmunity and allergy.
- Centre for Protein Engineering, Medical Research Council Centre, Hills Road, Cambridge CB2 2HQ, UK.
Organizational Affiliation: