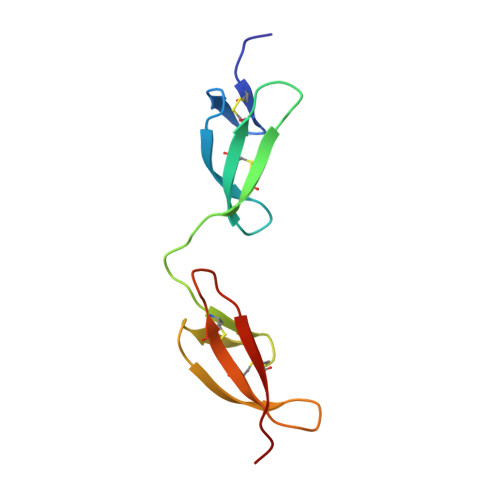
Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper.
Schwarz-Linek, U., Werner, J.M., Pickford, A.R., Gurusiddappa, S., Kim, J.H., Pilka, E.S., Briggs, J.A., Gough, T.S., Hook, M., Campbell, I.D., Potts, J.R.(2003) Nature 423: 177-181
- PubMed: 12736686
- DOI: https://doi.org/10.1038/nature01589
- Primary Citation of Related Structures:
1O9A - PubMed Abstract:
Staphylococcus aureus and Streptococcus pyogenes, two important human pathogens, target host fibronectin (Fn) in their adhesion to and invasion of host cells. Fibronectin-binding proteins (FnBPs), anchored in the bacterial cell wall, have multiple Fn-binding repeats in an unfolded region of the protein. The bacterium-binding site in the amino-terminal domain (1-5F1) of Fn contains five sequential Fn type 1 (F1) modules. Here we show the structure of a streptococcal (S. dysgalactiae) FnBP peptide (B3) in complex with the module pair 1F12F1. This identifies 1F1- and 2F1-binding motifs in B3 that form additional antiparallel beta-strands on sequential F1 modules-the first example of a tandem beta-zipper. Sequence analyses of larger regions of FnBPs from S. pyogenes and S. aureus reveal a repeating pattern of F1-binding motifs that match the pattern of F1 modules in 1-5F1 of Fn. In the process of Fn-mediated invasion of host cells, therefore, the bacterial proteins seem to exploit the modular structure of Fn by forming extended tandem beta-zippers. This work is a vital step forward in explaining the full mechanism of the integrin-dependent FnBP-mediated invasion of host cells.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.
Organizational Affiliation: