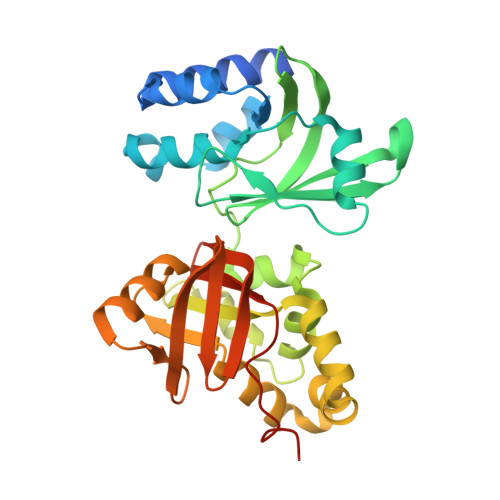
Novel Fold Revealed by the Structure of a Fas1 Domain Pair from the Insect Cell Adhesion Molecule Fasciclin I
Clout, N.J., Tisi, D., Hohenester, E.(2003) Structure 11: 197
- PubMed: 12575939
- DOI: https://doi.org/10.1016/s0969-2126(03)00002-9
- Primary Citation of Related Structures:
1O70 - PubMed Abstract:
Fasciclin I is an insect neural cell adhesion molecule consisting of four FAS1 domains, homologs of which are present in many bacterial, plant, and animal proteins. The crystal structure of FAS1 domains 3 and 4 of Drosophila fasciclin I reveals a novel domain fold, consisting of a seven-stranded beta wedge and a number of alpha helices. The two domains are arranged in a linear fashion and interact through a substantial polar interface. Missense mutations in the FAS1 domains of the human protein betaig-h3 cause corneal dystrophies. Many mutations alter highly conserved core residues, but the two most common mutations, affecting Arg-124 and Arg-555, map to exposed alpha-helical regions, suggesting reduced protein solubility as the disease mechanism.
- Biophysics Section, Department of Biological Sciences, Imperial College, SW7 2AZ, London, United Kingdom.
Organizational Affiliation: