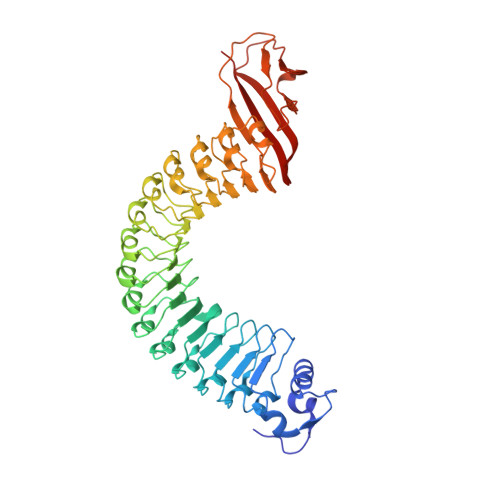
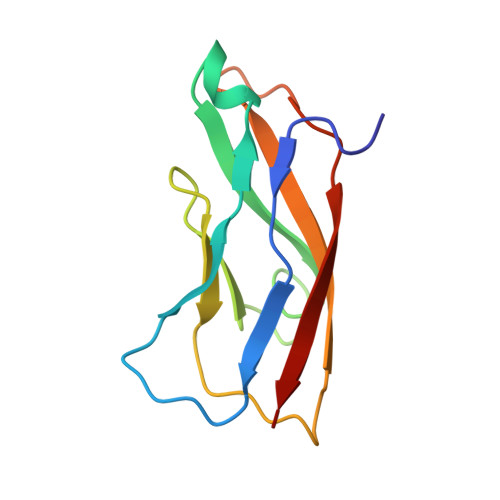
Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Schubert, W.-D., Urbanke, C., Ziehm, T., Beier, V., Machner, M.P., Domann, E., Wehland, J., Chakraborty, T., Heinz, D.W.(2002) Cell 111: 825
- PubMed: 12526809
- DOI: https://doi.org/10.1016/s0092-8674(02)01136-4
- Primary Citation of Related Structures:
1O6S, 1O6T, 1O6V - PubMed Abstract:
Listeria monocytogenes, a food-borne bacterial pathogen, enters mammalian cells by inducing its own phagocytosis. The listerial protein internalin (InlA) mediates bacterial adhesion and invasion of epithelial cells in the human intestine through specific interaction with its host cell receptor E-cadherin. We present the crystal structures of the functional domain of InlA alone and in a complex with the extracellular, N-terminal domain of human E-cadherin (hEC1). The leucine rich repeat (LRR) domain of InlA surrounds and specifically recognizes hEC1. Individual interactions were probed by mutagenesis and analytical ultracentrifugation. These include Pro16 of hEC1, a major determinant for human susceptibility to L. monocytogenes infection that is essential for intermolecular recognition. Our studies reveal the structural basis for host tro-pism of this bacterium and the molecular deception L. monocytogenes employs to exploit the E-cadherin system.
- Department of Structural Biology, German Research Center for Biotechnology (GBF), Mascheroder Weg 1, D-38124, Braunschweig, Germany.
Organizational Affiliation: