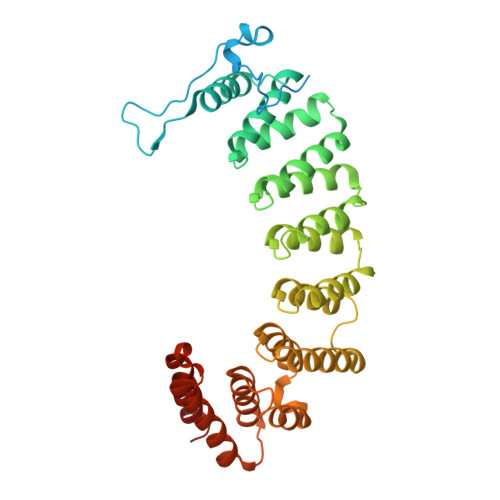
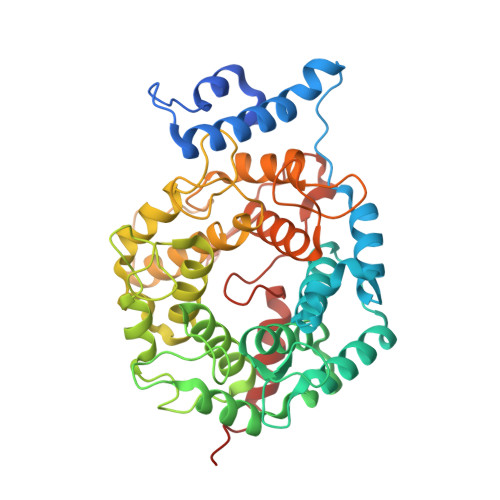
Biochemical and Structural Studies with Prenyl Diphosphate Analogues Provide Insights into Isoprenoid Recognition by Protein Farnesyl Transferase
Turek-Etienne, T.C., Strickland, C.L., Distefano, M.D.(2003) Biochemistry 42: 3716-3724
- PubMed: 12667062
- DOI: https://doi.org/10.1021/bi0266838
- Primary Citation of Related Structures:
1O1R, 1O1S, 1O1T - PubMed Abstract:
Protein farnesyl transferase (PFTase) catalyzes the reaction between farnesyl diphosphate and a protein substrate to form a thioether-linked prenylated protein. The fact that many prenylated proteins are involved in signaling processes has generated considerable interest in protein prenyl transferases as possible anticancer targets. While considerable progress has been made in understanding how prenyl transferases distinguish between related target proteins, the rules for isoprenoid discrimination by these enzymes are less well understood. To clarify how PFTase discriminates between FPP and larger prenyl diphosphates, we have examined the interactions between the enzyme and several isoprenoid analogues, GGPP, and the farnesylated peptide product using a combination of biochemical and structural methods. Two photoactive isoprenoid analogues were shown to inhibit yeast PFTase with K(I) values as low as 45 nM. Crystallographic analysis of one of these analogues bound to PFTase reveals that the diphosphate moiety and the two isoprene units bind in the same positions occupied by the corresponding atoms in FPP when bound to PFTase. However, the benzophenone group protrudes into the acceptor protein binding site and prevents the binding of the second (protein) substrate. Crystallographic analysis of geranylgeranyl diphosphate bound to PFTase shows that the terminal two isoprene units and diphosphate group of the molecule map to the corresponding atoms in FPP; however, the first and second isoprene units bulge away from the acceptor protein binding site. Comparison of the GGPP binding mode with the binding of the farnesylated peptide product suggests that the bulkier isoprenoid cannot rearrange to convert to product without unfavorable steric interactions with the acceptor protein. Taken together, these data do not support the "molecular ruler hypotheses". Instead, we propose a "second site exclusion model" in which PFTase binds larger isoprenoids in a fashion that prevents the subsequent productive binding of the acceptor protein or its conversion to product.
- Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, USA.
Organizational Affiliation: