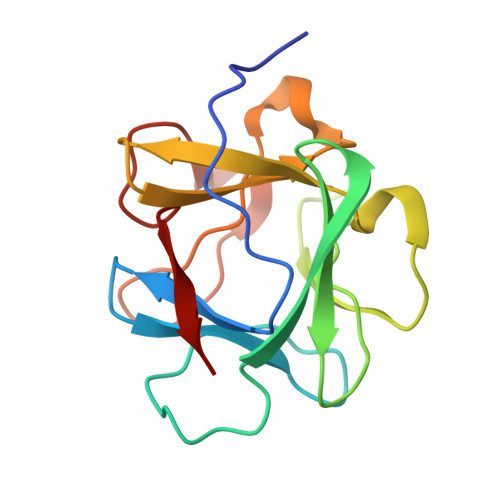
Accommodation of a highly symmetric core within a symmetric protein superfold.
Brych, S.R., Kim, J., Logan, T.M., Blaber, M.(2003) Protein Sci 12: 2704-2718
- PubMed: 14627732
- DOI: https://doi.org/10.1110/ps.03374903
- Primary Citation of Related Structures:
1JY0, 1M16, 1NZK, 1P63 - PubMed Abstract:
An alternative core packing group, involving a set of five positions, has been introduced into human acidic FGF-1. This alternative group was designed so as to constrain the primary structure within the core region to the same threefold symmetry present in the tertiary structure of the protein fold (the beta-trefoil superfold). The alternative core is essentially indistinguishable from the WT core with regard to structure, stability, and folding kinetics. The results show that the beta-trefoil superfold is compatible with a threefold symmetric constraint on the core region, as might be the case if the superfold arose as a result of gene duplication/fusion events. Furthermore, this new core arrangement can form the basis of a structural "building block" that can greatly simplify the de novo design of beta-trefoil proteins by using symmetric structural complementarity. Remaining asymmetry within the core appears to be related to asymmetry in the tertiary structure associated with receptor and heparin binding functionality of the growth factor.
- Kasha Laboratory, Institute of Molecular Biophysics and Department of Chemistry and Biochemistry, Florida State University, Tallahassee, Florida 32306-4380, USA.
Organizational Affiliation: