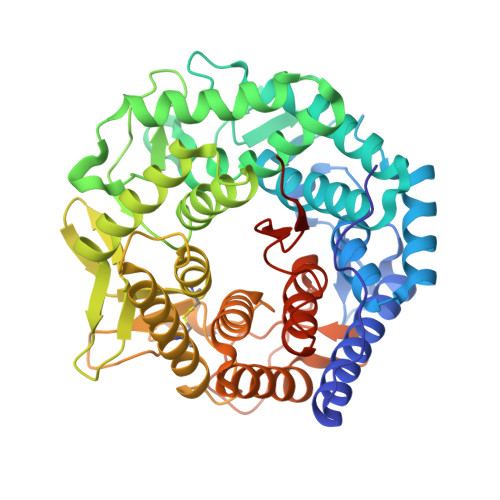
Structure of mouse Golgi alpha-mannosidase IA reveals the molecular basis for substrate specificity among class 1 (family 47 glycosylhydrolase) alpha1,2-mannosidases
Tempel, W., Karaveg, K., Liu, Z.-J., Rose, J., Wang, B.-C., Moremen, K.W.(2004) J Biological Chem 279: 29774-29786
- PubMed: 15102839
- DOI: https://doi.org/10.1074/jbc.M403065200
- Primary Citation of Related Structures:
1NXC - PubMed Abstract:
Three subfamilies of mammalian Class 1 processing alpha1,2-mannosidases (family 47 glycosidases) play critical roles in the maturation of Asn-linked glycoproteins in the endoplasmic reticulum (ER) and Golgi complex as well as influencing the timing and recognition for disposal of terminally unfolded proteins by ER-associated degradation. In an effort to define the structural basis for substrate recognition among Class 1 mannosidases, we have crystallized murine Golgi mannosidase IA (space group P2(1)2(1)2(1)), and the structure was solved to 1.5-A resolution by molecular replacement. The enzyme assumes an (alphaalpha)(7) barrel structure with a Ca(2+) ion coordinated at the base of the barrel similar to other Class 1 mannosidases. Critical residues within the barrel structure that coordinate the Ca(2+) ion or presumably bind and catalyze the hydrolysis of the glycone are also highly conserved. A Man(6)GlcNAc(2) oligosaccharide attached to Asn(515) in the murine enzyme was found to extend into the active site of an adjoining protein unit in the crystal lattice in a presumed enzyme-product complex. In contrast to an analogous complex previously isolated for Saccharomyces cerevisiae ER mannosidase I, the oligosaccharide in the active site of the murine Golgi enzyme assumes a different conformation to present an alternate oligosaccharide branch into the active site pocket. A comparison of the observed protein-carbohydrate interactions for the murine Golgi enzyme with the binding cleft topologies of the other family 47 glycosidases provides a framework for understanding the structural basis for substrate recognition among this class of enzymes.
- Department of Biochemistry and Molecular Biology, University of Georgia, Athens, Georgia 30602, USA.
Organizational Affiliation: