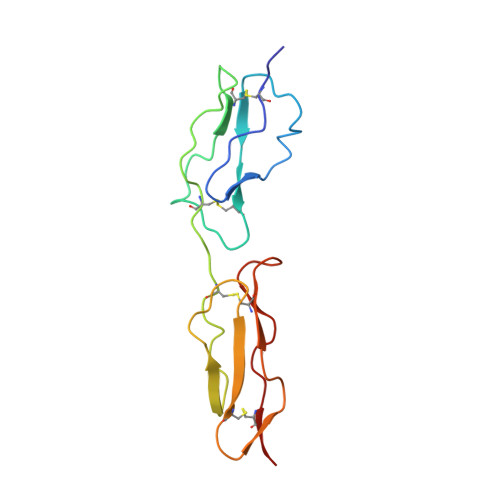
Solution structure of a functionally active fragment of decay-accelerating factor
Uhrinova, S., Lin, F., Ball, G., Bromek, K., Uhrin, D., Medof, M.E., Barlow, P.N.(2003) Proc Natl Acad Sci U S A 100: 4718-4723
- PubMed: 12672958
- DOI: https://doi.org/10.1073/pnas.0730844100
- Primary Citation of Related Structures:
1NWV - PubMed Abstract:
The second and third modules of human decay accelerating factor (DAF) are necessary and sufficient to accelerate decay of the classical pathway (CP) convertase of complement. No structure of a mammalian protein with decay-accelerating activity has been available to date. We therefore determined the solution structure of DAF modules 2 and 3 (DAF approximately 2,3). Structure-guided analysis of 24 mutants identified likely contact points between DAF and the CP convertase. Three (R96, R69, and a residue in the vicinity of L171) lie on DAF approximately 2,3's concave face. A fourth, consisting of K127 and nearby R100, is on the opposite face. Regions of module 3 remote from the semiflexible 2-3 interface seem not to be involved in binding to the CP convertase. DAF thus seems to occupy a groove on the CP convertase such that both faces of DAF close to the 2-3 junction (including a positively charged region that encircles the protein at this point) interact simultaneously. Alternative pathway convertase interactions with DAF require additional regions of CCP 3 lying away from the 2-3 interface, consistent with the established additional requirement of module 4 for alternative pathway regulation.
- Edinburgh Protein Interaction Centre, University of Edinburgh, Edinburgh EH9 3JJ, Scotland.
Organizational Affiliation: