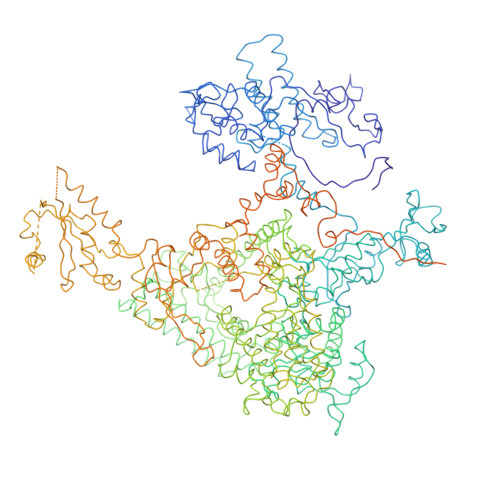
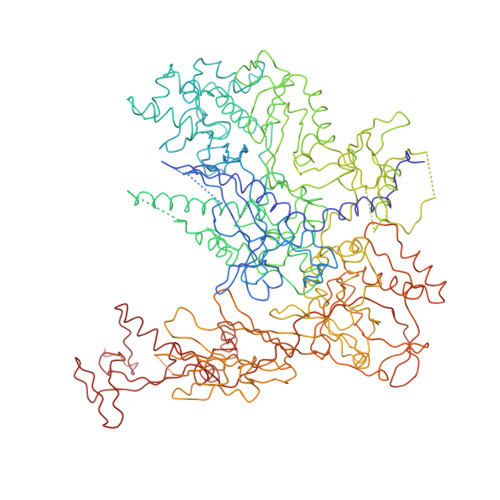
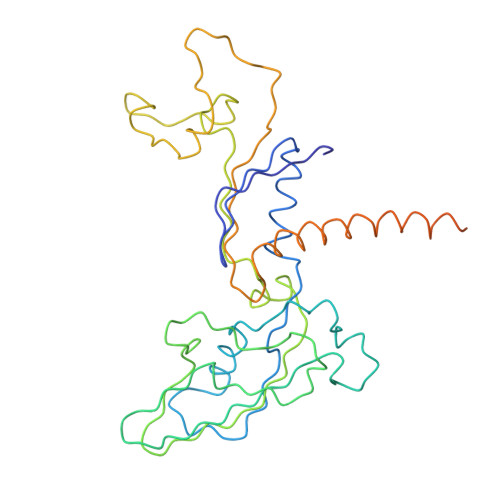
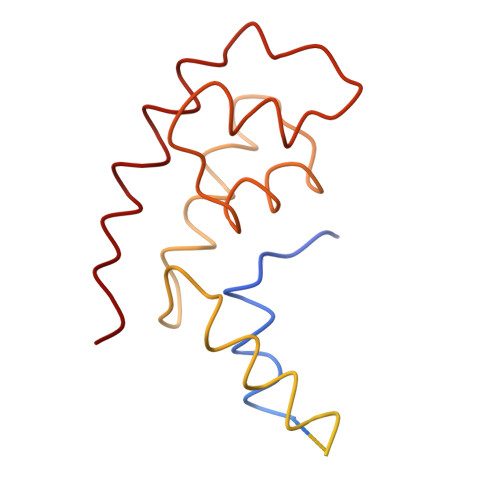
Architecture of initiation-competent 12-subunit RNA polymerase II
Armache, K.-J., Kettenberger, H., Cramer, P.(2003) Proc Natl Acad Sci U S A 100: 6964-6968
- PubMed: 12746495
- DOI: https://doi.org/10.1073/pnas.1030608100
- Primary Citation of Related Structures:
1NT9 - PubMed Abstract:
RNA polymerase (Pol) II consists of a 10-polypeptide catalytic core and the two-subunit Rpb4/7 complex that is required for transcription initiation. Previous structures of the Pol II core revealed a "clamp," which binds the DNA template strand via three "switch regions," and a flexible "linker" to the C-terminal repeat domain (CTD). Here we derived a model of the complete Pol II by fitting structures of the core and Rpb4/7 to a 4.2-A crystallographic electron density map. Rpb4/7 protrudes from the polymerase "upstream face," on which initiation factors assemble for promoter DNA loading. Rpb7 forms a wedge between the clamp and the linker, restricting the clamp to a closed position. The wedge allosterically prevents entry of the promoter DNA duplex into the active center cleft and induces in two switch regions a conformation poised for template-strand binding. Interaction of Rpb4/7 with the linker explains Rpb4-mediated recruitment of the CTD phosphatase to the CTD during Pol II recycling. The core-Rpb7 interaction and some functions of Rpb4/7 are apparently conserved in all eukaryotic and archaeal RNA polymerases but not in the bacterial enzyme.
- Institute of Biochemistry and Gene Center, University of Munich, Feodor-Lynen-Strasse 25, 81377 Munich, Germany.
Organizational Affiliation: