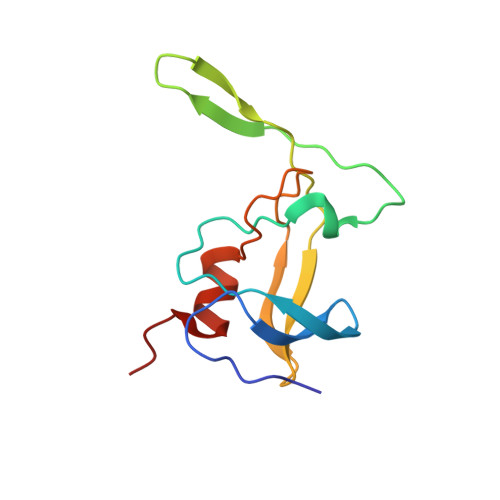
Three-dimensional structure of a monomeric form of a retroviral protease
Veverka, V., Bauerova, H., Zabransky, A., Lang, J., Ruml, T., Pichova, I., Hrabal, R.(2003) J Mol Biology 333: 771-780
- PubMed: 14568536
- DOI: https://doi.org/10.1016/j.jmb.2003.08.049
- Primary Citation of Related Structures:
1NSO - PubMed Abstract:
The assembly of Mason-Pfizer monkey virus Gag polyproteins into immature capsids and their cleavage by the encoded protease are temporally and spatially separated processes, making the virus a particularly useful model for investigation of protease activation. Here we present a high resolution NMR structure of a fully folded monomer of a 12 kDa M-PMV protease (wt 12 PR) and of a Cys7Ala/Asp26Asn/Cys106Ala mutant (12 PR(D26N/C7A/C106A)). The overall structures of both wt 12 PR and 12 PR(D26N/C7A/C106A) follow the conservative structural motif of other retroviral proteases. The most prominent difference from the canonical fold of retroviral proteases is the absence of the interfacial beta-sheet, which leads to the loss of the principal force stabilizing the dimer of M-PMV PR. The monomer-dimer equilibrium can be shifted in favor of the dimer by adding a substrate or an inhibitor, partially compensating for the missing role of the beta-sheet. We also show that cysteines C7 and C106 play a crucial role in stabilizing the dimer and consequently increasing the proteolytic activity of M-PMV PR. This is consistent with the role of reversible oxidative modification of the cysteine residues in the regulation of the maturation of assembled M-PMV capsids in the cytoplasm.
- NMR Laboratory, Institute of Chemical Technology in Prague, Technická, 5, Prague CZ-166 28, Czech Republic.
Organizational Affiliation: