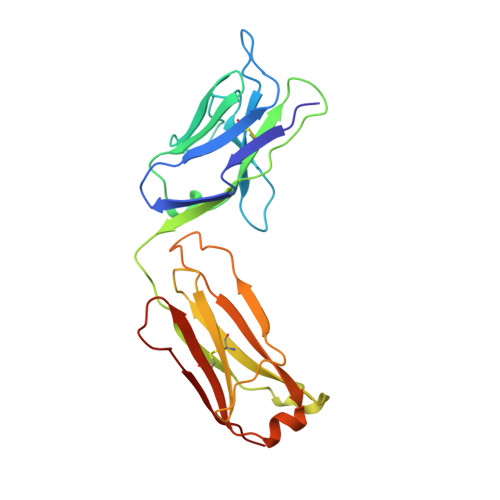
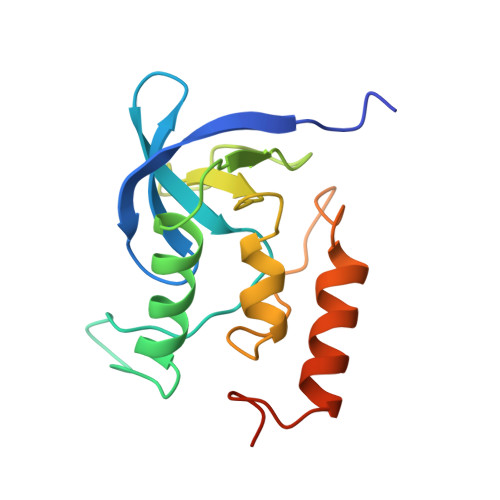
The crystal structure of the antibody N10-staphylococcal nuclease complex at 2.9 A resolution.
Bossart-Whitaker, P., Chang, C.Y., Novotny, J., Benjamin, D.C., Sheriff, S.(1995) J Mol Biology 253: 559-575
- PubMed: 7473734
- DOI: https://doi.org/10.1006/jmbi.1995.0573
- Primary Citation of Related Structures:
1NSN - PubMed Abstract:
The three-dimensional structure of the antibody N10 Fab fragment complexed with staphylococcal nuclease (SNase) has been determined to 2.9 A resolution. Eighteen residues from six complementarity-determining regions (CDR) recognize an epitope of five distinct SNase segments with a total of 17 residues. The overall shape of the antibody-antigen interface is U-shaped rather than the more or less rectangular interface seen in other antibody-protein antigen interfaces. Despite the U-shaped interface, the amount of surface buried in the complex, 828 A2 for SNase and 793 A2 for N10, is typical of antibody-protein antigen complexes. Contributing to the shape of the interface is the shortest antibody heavy chain-CDR3 loop reported to date, which probably allows access of bulk solvent in the center of the "U" interface. Another unusual feature of the N10 antibody is the 15 residue antibody light chain-CDR1, a length seen in only three other reported antibodies. Antibody light chain-CDR1 displays a previously unobserved conformation in its distal portion. Finally, although some of the movement observed in the antibody-bound SNase may be due to crystal contacts, it is clear that some side-chain rearrangements are the result of antigen-antibody interaction.
- Bristol-Myers Squibb Pharmaceutical Research Institute, Princeton, NJ 08543-4000, USA.
Organizational Affiliation: