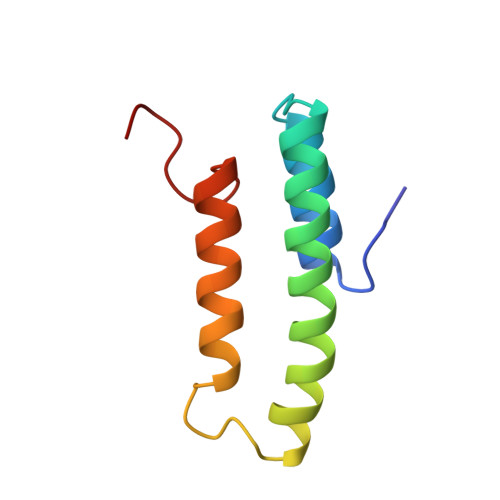
The solution structure of the N-terminal domain of alpha2-macroglobulin receptor-associated protein.
Nielsen, P.R., Ellgaard, L., Etzerodt, M., Thogersen, H.C., Poulsen, F.M.(1997) Proc Natl Acad Sci U S A 94: 7521-7525
- PubMed: 9207124
- DOI: https://doi.org/10.1073/pnas.94.14.7521
- Primary Citation of Related Structures:
1LRE, 1NRE - PubMed Abstract:
The three-dimensional structure of the N-terminal domain (residues 18-112) of alpha2-macroglobulin receptor-associated protein (RAP) has been determined by NMR spectroscopy. The structure consists of three helices composed of residues 23-34, 39-65, and 73-88. The three helices are arranged in an up-down-up antiparallel topology. The C-terminal 20 residues were shown not to be in a well defined conformation. A structural model for the binding of RAP to the family of low-density lipoprotein receptors is proposed. It defines a role in binding for both the unordered C terminus and the structural scaffold of the core structure. Pathogenic epitopes for the rat disease Heymann nephritis, an experimental model of human membranous glomerulonephritis, have been identified in RAP and in the large endocytic receptor gp330/megalin. Here we provide the three-dimensional structure of the pathogenic epitope in RAP. The amino acid residues known to form the epitope are in a helix-loop-helix conformation, and from the structure it is possible to rationalize the published results obtained from studies of fragments of the N-terminal domain.
- Carlsberg Laboratory, Department of Chemistry, Gamle Carlsberg Vej 10, DK-2500 Valby, Denmark.
Organizational Affiliation: