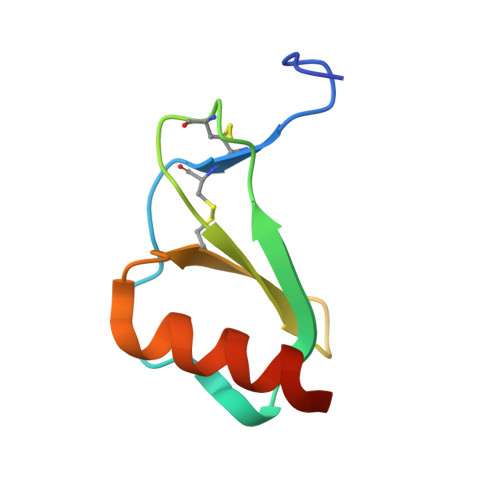
Structures of thymus and activation-regulated chemokine (TARC).
Asojo, O.A., Boulegue, C., Hoover, D.M., Lu, W., Lubkowski, J.(2003) Acta Crystallogr D Biol Crystallogr 59: 1165-1173
- PubMed: 12832759
- DOI: https://doi.org/10.1107/s0907444903009454
- Primary Citation of Related Structures:
1NR2, 1NR4 - PubMed Abstract:
Thymus and activation-regulated chemokine (TARC) is a CC chemokine that is mainly expressed in the thymus. TARC interacts primarily with the CCR4 receptor and to a lesser extent with the CCR8 receptor. The structures of TARC have been solved by molecular replacement in two space groups, triclinic (P1) and tetragonal (P4(1)), and refined to resolutions of 1.72 and 2.1 A, respectively, with R factors of 19.8% (R(free) = 24.1%) and 19.8% (R(free) = 27.7%), respectively. The search model originated from the crystal structure of another chemokine, RANTES, and proved to be only modestly similar to the refined structure of TARC. Whereas the tetragonal structure was easily solved using the program AMoRe, solution of the triclinic structure proved to be quite challenging and was obtained by combining the results from four different molecular-replacement programs (AMoRe, CNS, BEAST and EPMR), with subsequent extension of the gathered information. The tertiary structure of TARC is similar to that of other CC chemokines, with a three-stranded antiparallel beta-sheet flanked by a C-terminal helix. Both quaternary structures consist of dimers, which in the triclinic crystals pack further into tetramers. The TARC dimers are similar to those observed previously in the crystal structures of both MCP-1 and RANTES.
- Macromolecular Crystallography Laboratory, National Cancer Institute at Frederick, Frederick, MD 21702, USA.
Organizational Affiliation: