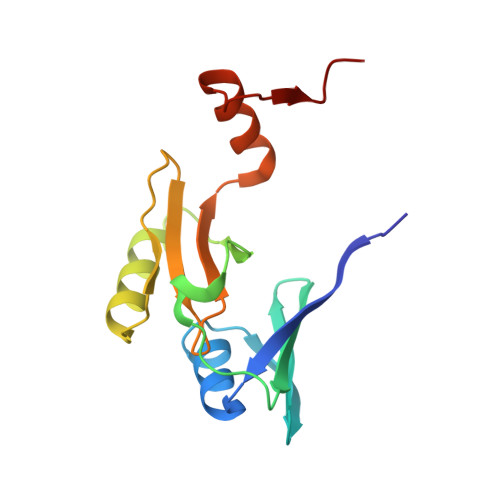
Structure of fosfomycin resistance protein FosA from transposon Tn2921.
Pakhomova, S., Rife, C.L., Armstrong, R.N., Newcomer, M.E.(2004) Protein Sci 13: 1260-1265
- PubMed: 15075406
- DOI: https://doi.org/10.1110/ps.03585004
- Primary Citation of Related Structures:
1NPB - PubMed Abstract:
The crystal structure of fosfomycin resistance protein FosA from transposon Tn2921 has been established at a resolution of 2.5 A. The protein crystallized without bound Mn(II) and K+, ions crucial for efficient catalysis, providing a structure of the apo enzyme. The protein maintains the three-dimensional domain-swapped arrangement of the paired betaalphabetabetabeta-motifs observed in the genomically encoded homologous enzyme from Pseudomonas aeruginosa (PA1129). The basic architecture of the active site is also maintained, despite the absence of the catalytically essential Mn(II). However, the absence of K+, which has been shown to enhance enzymatic activity, appears to contribute to conformational heterogeneity in the K(+)-binding loops.
- Departments of Biological Sciences and Chemistry, Louisiana State University, Baton Rouge, Louisiana 70803, USA. sveta@lsu.edu
Organizational Affiliation: