Structural Elucidation of the Specificity of the Antibacterial Agent Triclosan for Malarial Enoyl Acyl Carrier Protein Reductase
Perozzo, R., Kuo, M., Sidhu, A.S., Valiyaveettil, J.T., Bittman, R., Jacobs Jr., W.R., Fidock, D.A., Sacchettini, J.C.(2002) J Biological Chem 277: 13106-13114
- PubMed: 11792710
- DOI: https://doi.org/10.1074/jbc.M112000200
- Primary Citation of Related Structures:
1NHG, 1NHW, 1NNU, 1VRW - PubMed Abstract:
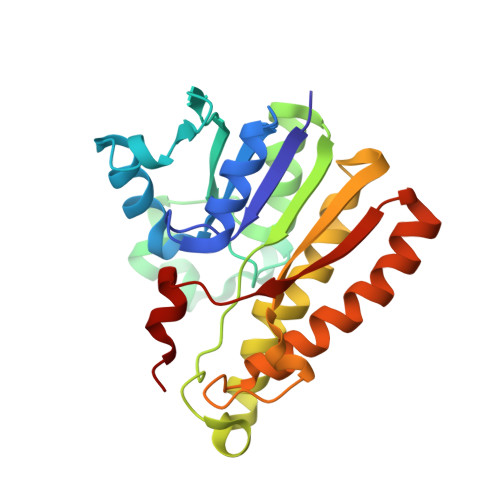
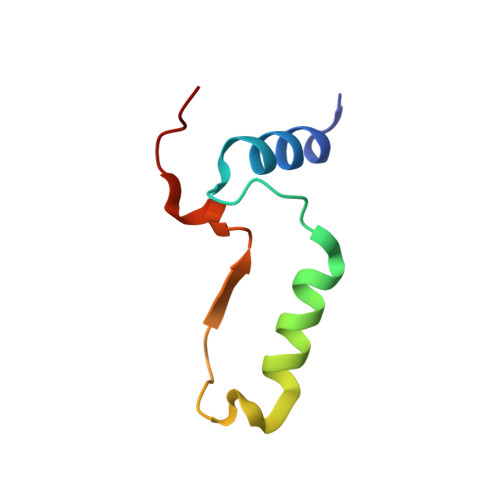
The human malaria parasite Plasmodium falciparum synthesizes fatty acids using a type II pathway that is absent in humans. The final step in fatty acid elongation is catalyzed by enoyl acyl carrier protein reductase, a validated antimicrobial drug target. Here, we report the cloning and expression of the P. falciparum enoyl acyl carrier protein reductase gene, which encodes a 50-kDa protein (PfENR) predicted to target to the unique parasite apicoplast. Purified PfENR was crystallized, and its structure resolved as a binary complex with NADH, a ternary complex with triclosan and NAD(+), and as ternary complexes bound to the triclosan analogs 1 and 2 with NADH. Novel structural features were identified in the PfENR binding loop region that most closely resembled bacterial homologs; elsewhere the protein was similar to ENR from the plant Brassica napus (root mean square for Calphas, 0.30 A). Triclosan and its analogs 1 and 2 killed multidrug-resistant strains of intra-erythrocytic P. falciparum parasites at sub to low micromolar concentrations in vitro. These data define the structural basis of triclosan binding to PfENR and will facilitate structure-based optimization of PfENR inhibitors.
- Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843-2128, USA.
Organizational Affiliation: