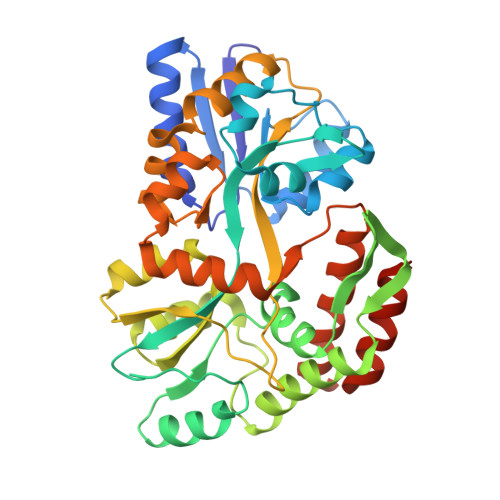
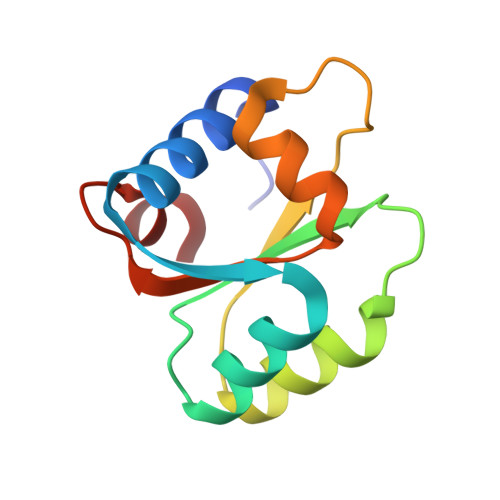
Inherent Protein Structural Flexibility at the RNA-binding Interface of L30e
Chao, J.A., Prasad, G.S., White, S.A., Stout, C.D., Williamson, J.R.(2003) J Mol Biology 326: 999-1004
- PubMed: 12589748
- DOI: https://doi.org/10.1016/s0022-2836(02)01476-6
- Primary Citation of Related Structures:
1NMU - PubMed Abstract:
The Saccharomyces cerevisiae ribosomal protein L30 autoregulates its own expression by binding to a purine-rich internal loop in its pre-mRNA and mRNA. NMR studies of L30 and its RNA complex showed that both the internal loop of the RNA as well as a region of the protein become substantially more ordered upon binding. A crystal structure of a maltose binding protein (MBP)-L30 fusion protein with two copies in the asymmetric unit has been determined. The flexible RNA-binding region in the L30 copies has two distinct conformations, one resembles the RNA bound form solved by NMR and the other is unique. Structure prediction algorithms also had difficulty accurately predicting this region, which is consistent with conformational flexibility seen in the NMR and X-ray crystallography studies. Inherent conformational flexibility may be a hallmark of regions involved in intermolecular interactions.
- Department of Molecular Biology, Department of Chemistry and The Skaggs Institute for Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: