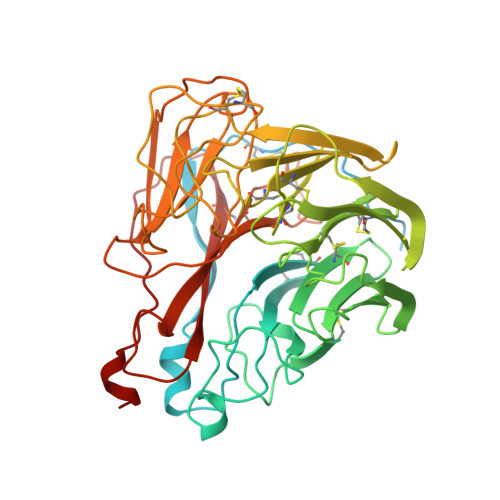
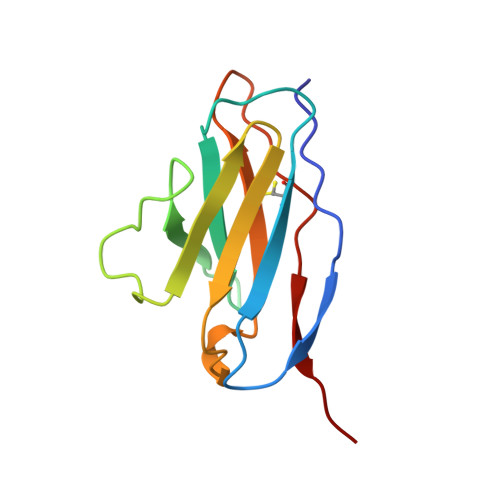
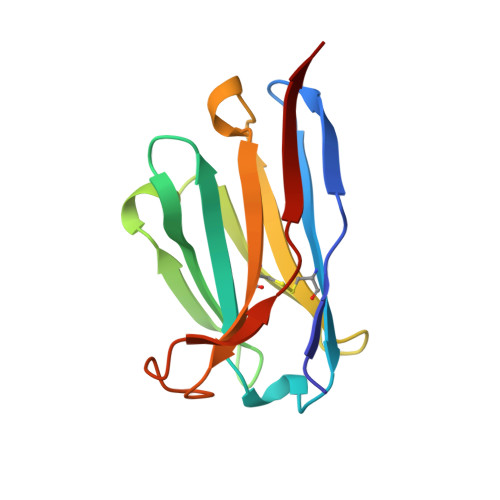
The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody
Malby, R.L., Tulip, W.R., Harley, V.R., McKimm-Breschkin, J.L., Laver, W.G., Webster, R.G., Colman, P.M.(1994) Structure 2: 733-746
- PubMed: 7994573
- DOI: https://doi.org/10.1016/s0969-2126(00)00074-5
- Primary Citation of Related Structures:
1NMB - PubMed Abstract:
While it is well known that different antibodies can be produced against a particular antigen, and even against a particular site on an antigen, up until now there have been no structural studies of cross-reacting antibodies of this type. One antibody-antigen complex whose structure is known is that of the influenza virus antigen, neuraminidase, in complex with the NC41 antibody. Another anti-neuraminidase antibody, NC10, binds to an overlapping site on the antigen. The structure of the complex formed by this antibody with neuraminidase is described here and compared with the NC41-containing complex. The crystal structure of the NC10 Fab-neuraminidase complex has been refined to a nominal resolution of 2.5A. Approximately 80% of the binding site of the NC10 antibody on neuraminidase overlaps with that of the NC41 antibody. The epitope residues of neuraminidase are often engaged in quite different interactions with the two antibodies. Although the NC10 and NC41 antibodies have identical amino acid sequences within the first complementarity determining region of their heavy chains, this is not the basis of the cross-reaction. The capacity of two different proteins to bind to the same target structure on a third protein need not be based on the existence of identical or homologous amino acid sequences within those proteins. As we have demonstrated, amino acid residues on the common target structure may be in quite different chemical environments, and may also adopt different conformations within two protein-protein complexes.
- Biomolecular Research Institute, Parkville, Victoria, Australia.
Organizational Affiliation: