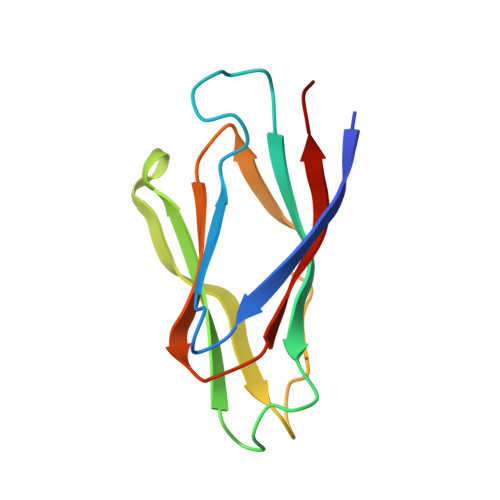
The crystal structure of Drosophila NLP-core Provides Insight into Pentamer Formation and Histone Binding
Namboodiri, V.M.H., Dutta, S., Akey, I.V., Head, J.F., Akey, C.W.(2003) Structure 11: 175-186
- PubMed: 12575937
- DOI: https://doi.org/10.1016/s0969-2126(03)00007-8
- Primary Citation of Related Structures:
1NLQ - PubMed Abstract:
The nucleoplasmin-like protein from Drosophila (dNLP) functions as a chaperone for core histones and may remodel chromatin in embryos. We now report the crystal structure of a dNLP-core pentamer at 1.5 A resolution. The monomer has an eight-stranded, beta barrel topology that is similar to nucleoplasmin (Np). However, a signature beta hairpin is tucked in along the lateral surface of the dNLP-core pentamer, while it extends outward in the Np-core decamer. Drosophila NLP and Np both assemble histone octamers. This process may require each chaperone to form a decamer, which would create symmetric binding sites for the histones. Conformational differences between dNLP and Np may reflect their different oligomeric states, while a conserved, nonpolar subunit interface may allow conformational plasticity during histone binding.
- Department of Physiology and Biophysics, Boston University School of Medicine, 700 Albany Street, Boston, MA 02118, USA.
Organizational Affiliation: