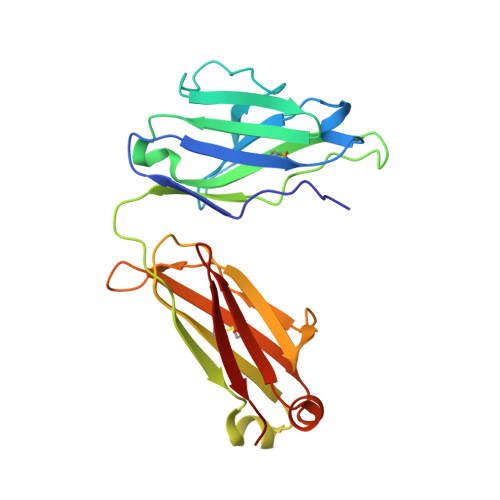
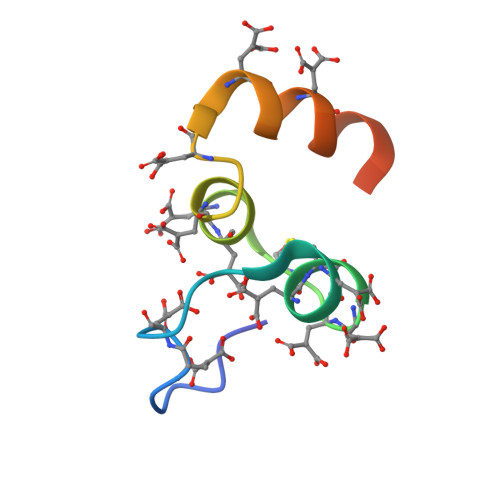
Crystal Structure of the Calcium-stabilized Human Factor IX Gla Domain Bound to a Conformation-specific Anti-factor IX Antibody.
Huang, M., Furie, B.C., Furie, B.(2004) J Biological Chem 279: 14338-14346
- PubMed: 14722079
- DOI: https://doi.org/10.1074/jbc.M314011200
- Primary Citation of Related Structures:
1NL0, 3D69 - PubMed Abstract:
The binding of Factor IX to membranes during blood coagulation is mediated by the N-terminal gamma-carboxyglutamic acid-rich (Gla) domain, a membrane-anchoring domain found on vitamin K-dependent blood coagulation and regulatory proteins. Conformation-specific anti-Factor IX antibodies are directed at the calcium-stabilized Gla domain and interfere with Factor IX-membrane interaction. One such antibody, 10C12, recognizes the calcium-stabilized form of the Gla domain of Factor IX. We prepared the fully carboxylated Gla domain of Factor IX by solid phase peptide synthesis and crystallized Factor IX-(1-47) in complex with Fab fragments of the 10C12 antibody. The overall structure of the Gla domain in the Factor IX-(1-47)-antibody complex at 2.2 A is similar to the structure of the Factor IX Gla domain in the presence of calcium ions as determined by NMR spectroscopy (Freedman, S. J., Furie, B. C., Furie, B., and Baleja, J. D. (1995) Biochemistry 34, 12126-12137) and by x-ray crystallography (Shikamoto, Y., Morita, T., Fujimoto, Z., and Mizuno, H. (2003) J. Biol. Chem. 278, 24090-24094). The complex structure shows that the complementarity determining region loops of the 10C12 antibody form a hydrophobic pocket to accommodate the hydrophobic patch of the Gla domain consisting of Leu-6, Phe-9, and Val-10. Polar interactions also play an important role in the antibody-antigen recognition. Furthermore, the calcium coordination network of the Factor IX Gla domain is different than in Gla domain structures of other vitamin K-dependent proteins. We conclude that this antibody is directed at the membrane binding site in the omega loop of Factor IX and blocks Factor IX function by inhibiting its interaction with membranes.
- Center for Hemostasis, Thrombosis and Vascular Biology, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, Massachusetts 02215, USA.
Organizational Affiliation: