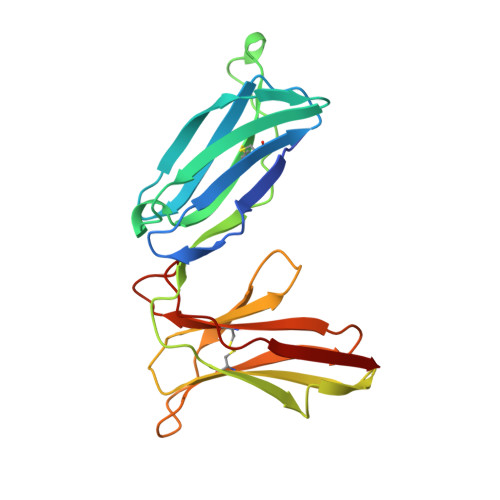
Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors.
Fan, Q.R., Mosyak, L., Winter, C.C., Wagtmann, N., Long, E.O., Wiley, D.C.(1997) Nature 389: 96-100
- PubMed: 9288975
- DOI: https://doi.org/10.1038/38028
- Primary Citation of Related Structures:
1NKR - PubMed Abstract:
Abnormal cells deficient in class I major histocompatibility complex (MHC) expression are lysed by a class of lymphocytes called natural killer (NK) cells. This lysis provides a defence against pathogens and tumour cells that downregulate MHC expression to avoid an MHC-restricted, T-cell immune response. Normal cells escape lysis because their MHC molecules are recognized by NK-cell inhibitory receptors, which inhibit lysis. Several such inhibitory receptor families have been described in humans and mice. In the human killer-cell inhibitory receptor family, individual p58 members are specific for a subset of class I human leukocyte antigen (HLA)-C molecules. The human p58 natural killer-cell inhibitory receptor clone 42 recognizes HLA-Cw4, -Cw2 and -Cw6, but not HLA-Cw3, -Cw2, -Cw7 or -Cw8, which are recognized by p58 killer-cell inhibitor receptor clone 43. We have determined the X-ray structure of the p58 NK-cell inhibitory receptor clone 42 at 1.7-A resolution. The structure has tandem immunoglobulin-like domains positioned at an acute, 60-degree angle. Loops on the outside of the elbow between the domains form a binding site projected away from the NK-cell surface. The topology of the domains and their arrangement relative to each other reveal a relationship to the haematopoietic receptor family, with implications for the signalling mechanism in NK cells.
- Department of Molecular and Cellular Biology, Howard Hughes Medical Institute, Harvard University, Cambridge, Massachusetts 02138, USA. fan@crystal.harvard.edu
Organizational Affiliation: