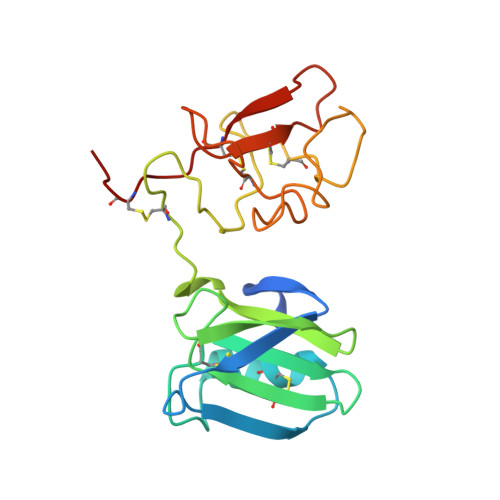
Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Chirgadze, D.Y., Hepple, J.P., Zhou, H., Byrd, R.A., Blundell, T.L., Gherardi, E.(1999) Nat Struct Biol 6: 72-79
- PubMed: 9886295
- DOI: https://doi.org/10.1038/4947
- Primary Citation of Related Structures:
1NK1 - PubMed Abstract:
Although ligand-induced receptor dimerization is a common prerequisite for receptor activation, the mode by which different growth factors bind their receptors and cause them to dimerize varies considerably. Here we report the crystal structure at 2.5 A resolution of NK1, a receptor-binding fragment and a natural splice variant of hepatocyte growth factor/scatter factor (HGF/SF). NK1 assembles as a homodimer in the asymmetric unit, revealing a novel mode of growth factor dimerization produced by close packing of the N domain of one subunit and the kringle domain of the other, thus bringing the two linkers in close proximity. The structure suggests the presence of a binding site for heparan sulfate chains and a mechanism by which the NK1 dimer may engage two receptor molecules through clusters of amino acids located on each protomer and on opposite surfaces of the homodimer. We also report that short (14-mer) heparin fragments effectively dimerize NK1 in solution, implying that heparan sulfate chains may stabilize the NK1 dimer. These results provide a basis for the agonistic activity of NK1 and have implications for the mechanism of receptor binding of HGF/SF.
- Department of Biochemistry, University of Cambridge, UK.
Organizational Affiliation: