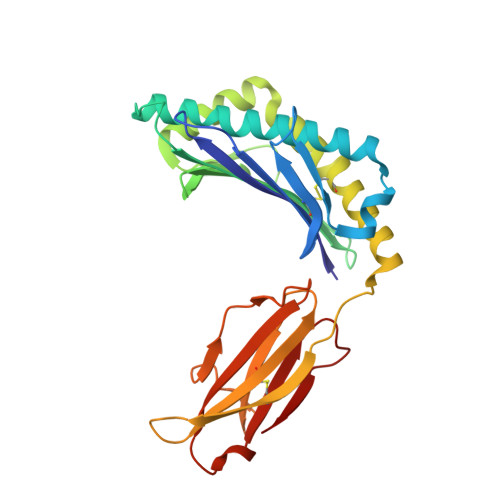
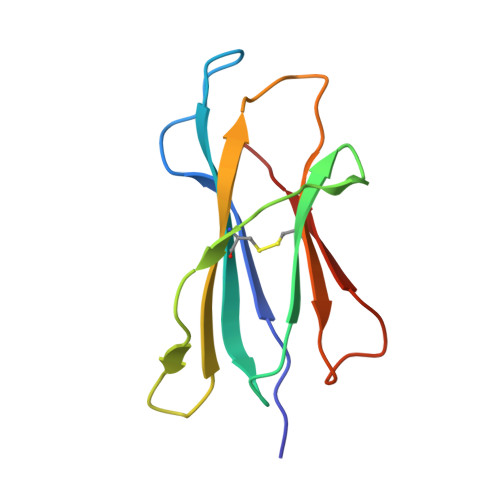
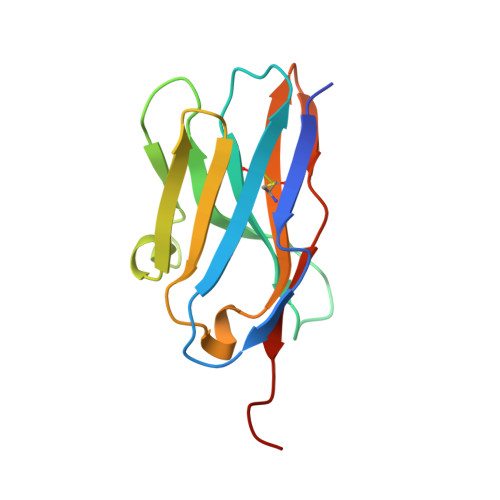
The Crystal Structure of a TL/CD8alphaalpha Complex at 2.1 A resolution: Implications for modulation of T cell activation and memory
Liu, Y., Xiong, Y., Naidenko, O.V., Liu, J.H., Zhang, R., Joachimiak, A., Kronenberg, M., Cheroutre, H., Reinherz, E.L., Wang, J.H.(2003) Immunity 18: 205-215
- PubMed: 12594948
- DOI: https://doi.org/10.1016/s1074-7613(03)00027-x
- Primary Citation of Related Structures:
1NEZ - PubMed Abstract:
TL is a nonclassical MHC class I molecule that modulates T cell activation through relatively high-affinity interaction with CD8alphaalpha. To investigate how the TL/CD8alphaalpha interaction influences TCR signaling, we characterized the structure of the TL/CD8alphaalpha complex using X-ray crystallography. Unlike antigen-presenting molecules, the TL antigen-binding groove is occluded by specific conformational changes. This feature eliminates antigen presentation, severely hampers direct TCR recognition, and prevents TL from participating in the TCR activation complex. At the same time, the TL/CD8alphaalpha interaction is strengthened through subtle structure changes in the TL alpha3 domain. Thus, TL functions to sequester and redirect CD8alphaalpha away from the TCR, modifying lck-dependent signaling.
- Laboratory of Immunobiology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: