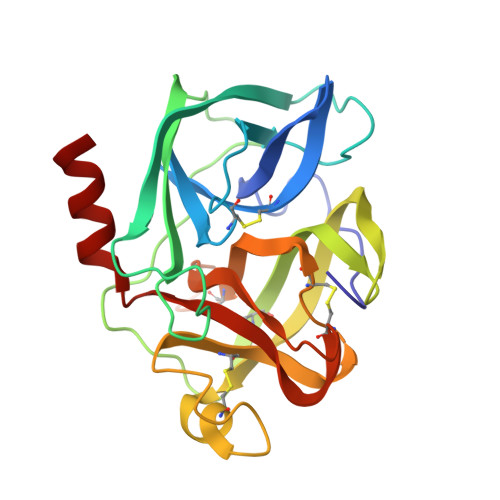
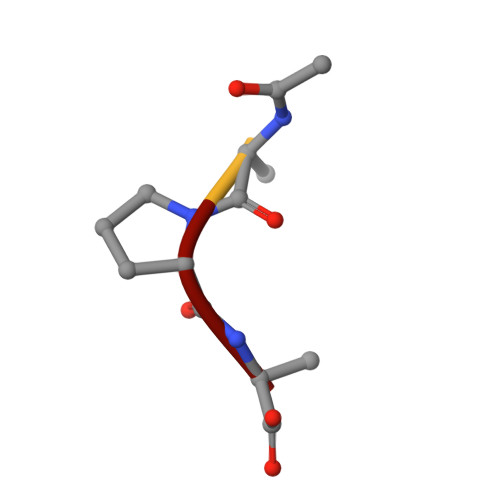
Structure of the product complex of acetyl-Ala-Pro-Ala with porcine pancreatic elastase at 1.65 A resolution.
Meyer Jr., E.F., Radhakrishnan, R., Cole, G.M., Presta, L.G.(1986) J Mol Biology 189: 533-539
- PubMed: 3640831
- DOI: https://doi.org/10.1016/0022-2836(86)90322-0
- Primary Citation of Related Structures:
1NES - PubMed Abstract:
A single crystal of porcine pancreatic elastase was mounted in a thin-walled capillary and allowed to react with acetyl-Ala-Pro-Ala-paranitroanalide. Diffraction data to 1.65 A resolution were measured and the isomorphous structure was solved from the difference Fourier map. The structure contains two surprises. Two molecules of the product: acetyl-Ala-Pro-Ala molecule are bound in the extended binding site. Both molecules are bound backwards with respect to the established mode of peptide binding.