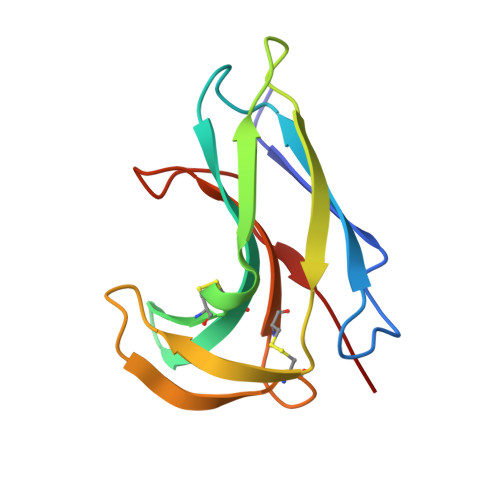
Crystal structure of neocarzinostatin, an antitumor protein-chromophore complex.
Kim, K.H., Kwon, B.M., Myers, A.G., Rees, D.C.(1993) Science 262: 1042-1046
- PubMed: 8235619
- DOI: https://doi.org/10.1126/science.8235619
- Primary Citation of Related Structures:
1NCO - PubMed Abstract:
Structures of the protein-chromophore complex and the apoprotein form of neocarzinostatin were determined at 1.8 angstrom resolution. Neocarzinostatin is composed of a labile chromophore with DNA-cleaving activity and a stabilizing protein. The chromophore displays marked nonlinearity of the triple bonds and is bound noncovalently in a pocket formed by the two protein domains. The chromophore pi-face interacts with the phenyl ring edges of Phe52 and Phe78. The amino sugar and carbonate groups of the chromophore are solvent exposed, whereas the epoxide, acetylene groups, and carbon C-12, the site of nucleophilic thiol addition during chromophore activation, are unexposed. The position of the amino group of the chromophore carbohydrate relative to C-12 supports the idea that the amino group plays a role in thiol activation.
- Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena 91125.
Organizational Affiliation: