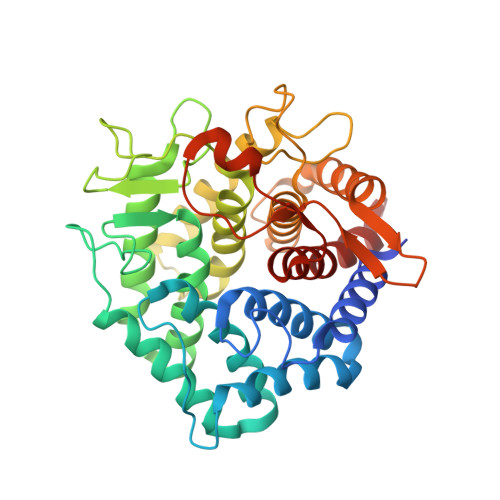
1.6 A crystal structure of YteR protein from Bacillus subtilis, a predicted lyase.
Zhang, R., Minh, T., Lezondra, L., Korolev, S., Moy, S.F., Collart, F., Joachimiak, A.(2005) Proteins 60: 561-565
- PubMed: 15906318
- DOI: https://doi.org/10.1002/prot.20410
- Primary Citation of Related Structures:
1NC5 - Structural Biology Center, Midwest Center for Structural Genomics, Biosciences, Argonne National Laboratory, 9700 South Cass Ave., Bldg. 202, Argonne, IL 60439, USA.
Organizational Affiliation: