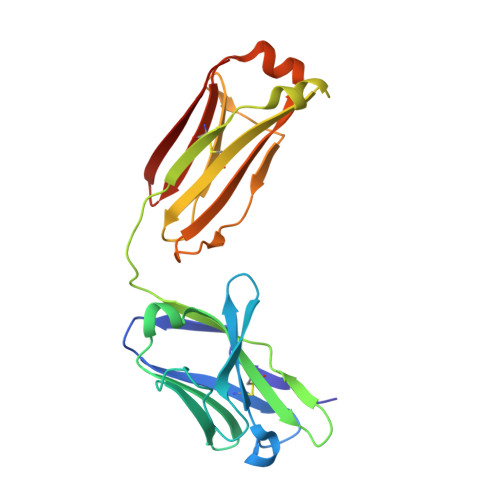
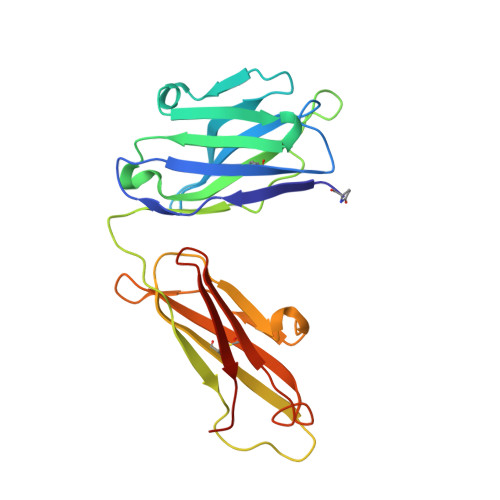
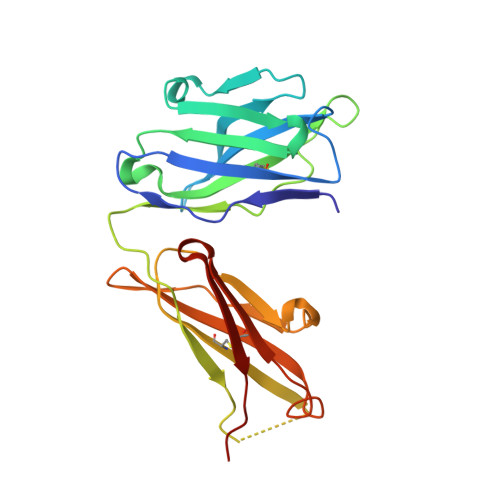
Crystal structures of two complexes of the rare-earth-DOTA-binding antibody 2D12.5: ligand generality from a chiral system.
Corneillie, T.M., Fisher, A.J., Meares, C.F.(2003) J Am Chem Soc 125: 15039-15048
- PubMed: 14653738
- DOI: https://doi.org/10.1021/ja037236y
- Primary Citation of Related Structures:
1NC2, 1NC4 - PubMed Abstract:
We report the crystal structures of antibody 2D12.5 Fab bound to an yttrium-DOTA analogue and separately to a gadolinium-DOTA analogue. The rare earth elements have many useful properties as probes, and 2D12.5 binds the DOTA (1,4,7,10-tetraazacyclododecane-N,N',N' ',N' "-tetraacetic acid) complexes of all of them (Corneillie et al. J. Am. Chem. Soc. 2003, 125, 3436-3437). The structures show that there are no direct protein-metal interactions: a bridging water acts as a link between the protein and metal, with the chelate present as the M isomer in each case. DOTA forms an amphipathic cylinder with the charged carboxylate groups toward the face of the chelate near the metal ion, while nonpolar methylene groups from the macrocycle and the carboxymethyl groups occupy the rear and sides of the molecule. The orientation of the metal-DOTA in the 2D12.5 complex places most of the methylene carbon atoms of DOTA in hydrophobic contact with aromatic protein side chains. Other binding interactions between the metal complex and the antibody include a bidentate salt bridge, four direct H-bonds, and four to five water-mediated H-bonds. We find that 2D12.5 exhibits enantiomeric binding generality, binding yttrium chelates in both Lambda(deltadeltadeltadelta) and Delta(lambdalambdalambdalambda) configurations with modestly different affinities. This develops from the symmetrical nature of the DOTA chelate, which places heteroatoms and hydrophobic atoms in approximately the same relative positions regardless of the helicity of DOTA.
- Department of Chemistry, University of California, One Shields Avenue, Davis, California 95616, USA.
Organizational Affiliation: