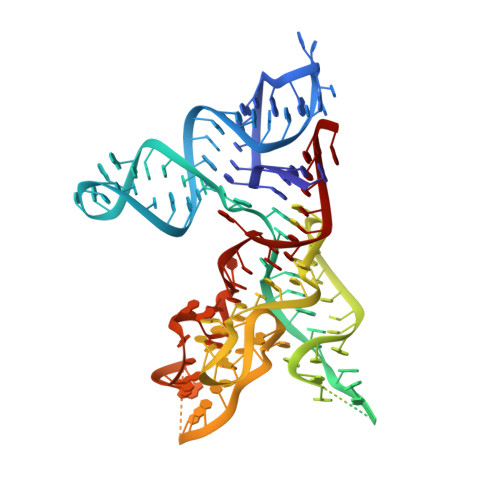
Crystal structure of the specificity domain of Ribonuclease P
Krasilnikov, A.S., Yang, X., Pan, T., Mondragon, A.(2003) Nature 421: 760-764
- PubMed: 12610630
- DOI: https://doi.org/10.1038/nature01386
- Primary Citation of Related Structures:
1NBS - PubMed Abstract:
RNase P is the only endonuclease responsible for processing the 5' end of transfer RNA by cleaving a precursor and leading to tRNA maturation. It contains an RNA component and a protein component and has been identified in all organisms. It was one of the first catalytic RNAs identified and the first that acts as a multiple-turnover enzyme in vivo. RNase P and the ribosome are so far the only two ribozymes known to be conserved in all kingdoms of life. The RNA component of bacterial RNase P can catalyse pre-tRNA cleavage in the absence of the RNase P protein in vitro and consists of two domains: a specificity domain and a catalytic domain. Here we report a 3.15-A resolution crystal structure of the 154-nucleotide specificity domain of Bacillus subtilis RNase P. The structure reveals the architecture of this domain, the interactions that maintain the overall fold of the molecule, a large non-helical but well-structured module that is conserved in all RNase P RNA, and the regions that are involved in interactions with the substrate.
- Department of Biochemistry, Molecular Biology and Cell Biology, Northwestern University, Evanston, Illinois 60208, USA.
Organizational Affiliation: