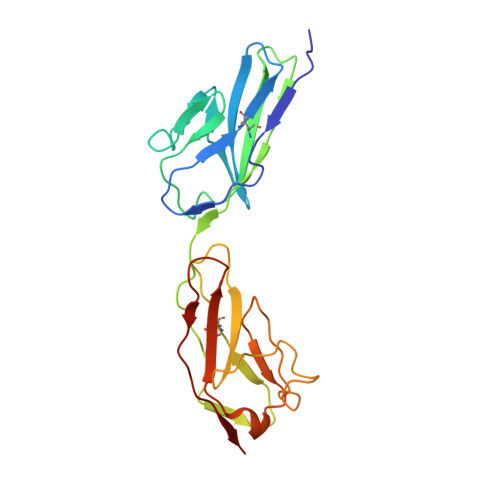
Crystal structure of human junctional adhesion molecule 1: Implications for reovirus binding
Prota, A.E., Campbell, J.A., Schelling, P., Forrest, J.C., Watson, M.J., Peters, T.R., Aurrand-Lions, M., Imhof, B.A., Dermody, T.S., Stehle, T.(2003) Proc Natl Acad Sci U S A 100: 5366-5371
- PubMed: 12697893
- DOI: https://doi.org/10.1073/pnas.0937718100
- Primary Citation of Related Structures:
1NBQ - PubMed Abstract:
Reovirus attachment to cells is mediated by the binding of viral attachment protein sigma 1 to junctional adhesion molecule 1 (JAM1). The crystal structure of the extracellular region of human JAM1 (hJAM1) reveals two concatenated Ig-type domains with a pronounced bend at the domain interface. Two hJAM1 molecules form a dimer that is stabilized by extensive ionic and hydrophobic contacts between the N-terminal domains. This dimeric arrangement is similar to that observed previously in the murine homolog of JAM1, indicating physiologic relevance. However, differences in the dimeric structures of hJAM1 and murine JAM1 suggest that the interface is dynamic, perhaps as a result of its ionic nature. We demonstrate that hJAM1, but not the related proteins hJAM2 and hJAM3, serves as a reovirus receptor, which provides insight into sites in hJAM1 that likely interact with sigma 1. In addition, we present evidence that the previously reported structural homology between sigma 1 and the adenovirus attachment protein, fiber, also extends to their respective receptors, which form similar dimeric structures. Because both receptors are located at regions of cell-cell contact, this similarity suggests that reovirus and adenovirus use conserved mechanisms of entry and pathways of infection.
- Laboratory of Developmental Immunology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Organizational Affiliation: