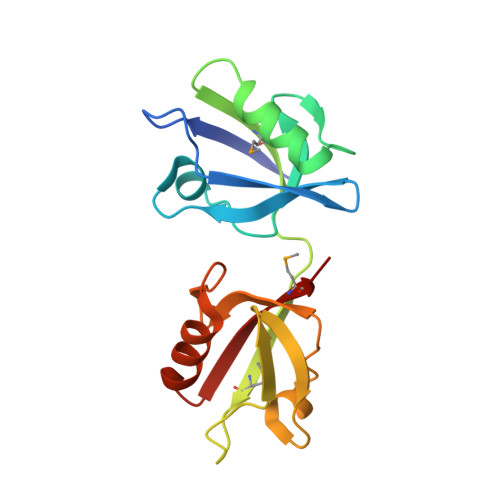
PDZ Tandem of Human Syntenin: Crystal Structure and Functional Properties
Kang, B.S., Cooper, D.R., Jelen, F., Devedjiev, Y., Derewenda, U., Dauter, Z., Otlewski, J., Derewenda, Z.S.(2003) Structure 11: 459-468
- PubMed: 12679023
- DOI: https://doi.org/10.1016/s0969-2126(03)00052-2
- Primary Citation of Related Structures:
1N99 - PubMed Abstract:
Syntenin, a 33 kDa protein, interacts with several cell membrane receptors and with merlin, the product of the causal gene for neurofibromatosis type II. We report a crystal structure of the functional fragment of human syntenin containing two canonical PDZ domains, as well as binding studies for full-length syntenin, the PDZ tandem, and isolated PDZ domains. We show that the functional properties of syntenin are a result of independent interactions with target peptides, and that each domain is able to bind peptides belonging to two different classes: PDZ1 binds peptides from classes I and III, while PDZ2 interacts with classes I and II. The independent binding of merlin by PDZ1 and syndecan-4 by PDZ2 provides direct evidence for the coupling of syndecan-mediated signaling to actin regulation by merlin.
- Department of Molecular Physiology and Biological Physics and The Cancer Center, University of Virginia, Charlottesville, VA 22908, USA.
Organizational Affiliation: