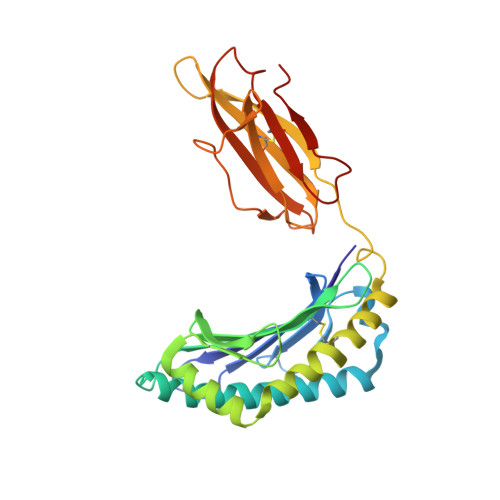
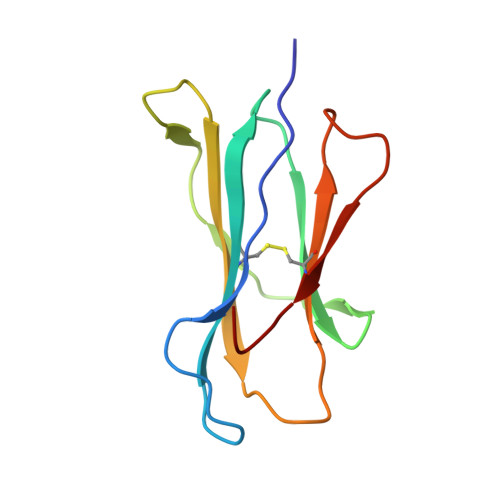
Structural analysis of mycobacterial and murine hsp60 epitopes in complex with the class I MHC molecule H-2D(b)
Ciatto, C., Capitani, G., Tissot, A.C., Pecorari, F., Plueckthun, A., Gruetter, M.G.(2003) FEBS Lett 543: 11-15
- PubMed: 12753896
- DOI: https://doi.org/10.1016/s0014-5793(03)00325-9
- Primary Citation of Related Structures:
1N3N - PubMed Abstract:
The decameric peptide SALQNAASIA from the Mycobacterium bovis heat shock protein (hsp) 60 is recognized by the murine T-cell receptor UZ-3-4 in complex with the murine class I major histocompatibility complex molecule H-2D(b). This T-cell receptor cross-reacts with the H-2D(b)-bound non-homologous decameric peptide KDIGNIISDA from the murine hsp60, but does not recognize the nonameric mycobacterial peptide SALQNAASI. Cross-recognition of the KDIGNIISDA/H-2D(b) complex induces autoimmune pathology in immunodeficient mice. We solved the X-ray crystal structure of the SALQNAASIA/H-2D(b) complex at 3.0 A resolution, and we modelled the KDIGNIISDA and SALQNAASI peptides in the H-2D(b) binding site. The structural analysis of the H-2D(b)-bound hsp60 epitopes offers insight into T-cell receptor cross-reactivity.
- Department of Biochemistry, University of Zürich, Winterthurerstrasse 190, CH-8057, Zürich, Switzerland.
Organizational Affiliation: